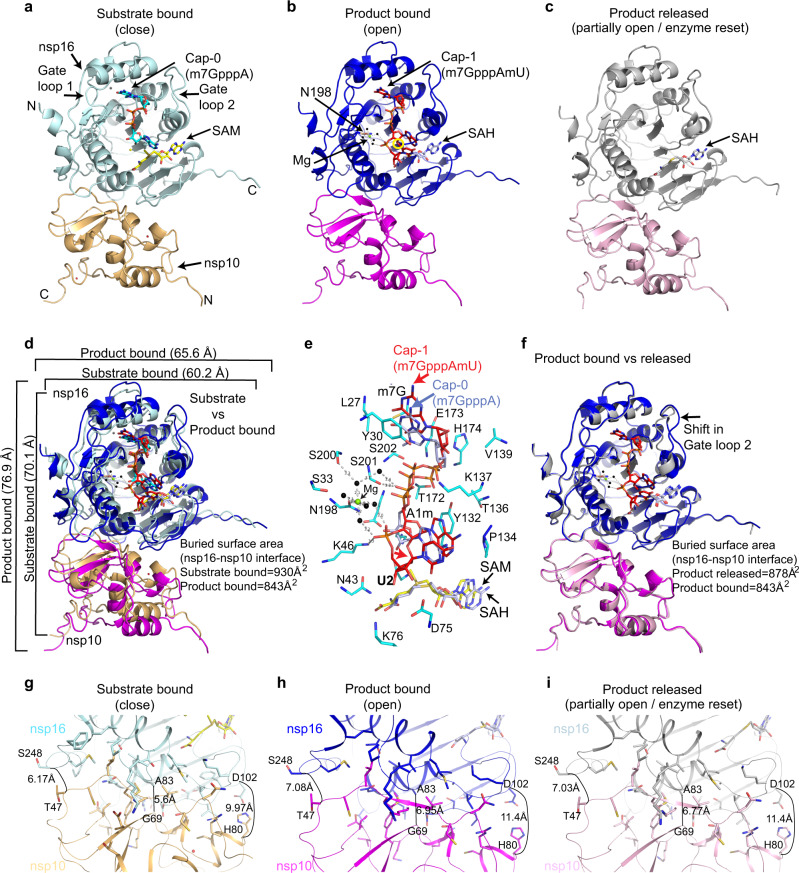
Fig. 1. Structures of SARS-CoV-2 nsp16/nsp10 complexes.
a The substrate (me7GpppA, cyan stick) and methyl donor S-adenosyl-l-methionine (SAM, yellow stick)-bound nsp16 (cyan)/nsp10 (orange) complex (PDB ID, 6WKS)9 represent a closed form. b The product (me7GpppAmU, red stick; byproduct S-adenosyl homocysteine [SAH, gray stick])-bound nsp16 (blue)/nsp10 (magenta) in an open state. A yellow circle shows the methylated ribose (2’-O-me) of N1 (A) base. c The SAH (gray)-bound nsp16 (gray)/nsp10 (pink) represents a partially open or enzyme reset state. d Secondary structure-based overlay of nsp16 in substrate- and product-bound states clearly shows the universal expansion of the enzyme upon 2’-O methylation. e A close-up view of Cap-1-binding and catalytic pocket of the product structure shows nsp16 residues (cyan sticks) interacting with Cap-1 (red). A positional change in orientation of the substrate (Cap-0, blue) from the “closed” structure determined previously9 is shown. f An overlay of the product (Cap-1)- and byproduct (SAH)-bound structures shows change in the orientation of gate loop 2. Reduction in buried surface area between nsp16/nsp10 in fully and partially open structures (compared to substrate-bound closed state) is shown (g–i).