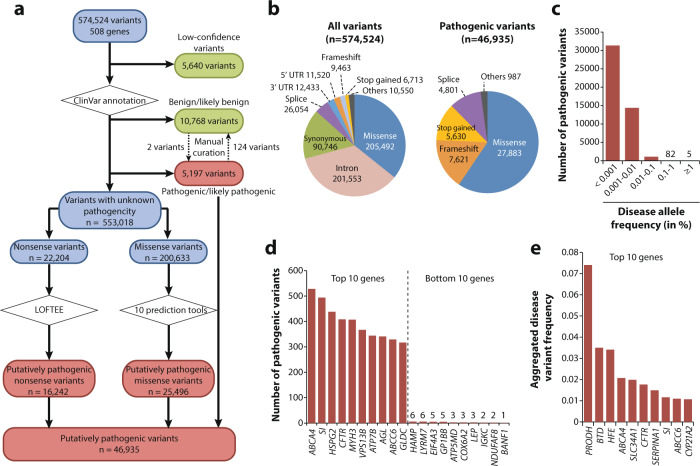
Fig. 1. The landscape of pathogenic variability differs drastically between genes.
a Algorithm for pathogenic variant selection in human autosomal recessive disease genes. In total, 574,524 variants were identified in 508 genes associated with autosomal recessive disease. In a first step, we removed low-confidence and benign variants, whereas all variants in these genes that were known to be pathogenic according to ClinVar (n = 5197) were included for further analyses. Rare variants with unclear pathogenicity were analyzed computationally using LOFTEE for nonsense variants or ten computational prediction tools for missense variants (see “Methods” section for details). Variants that were deemed as pathogenic by LOFTEE (n = 16,242) or all missense prediction algorithms (n = 25,496) were combined with the variants with pathogenic ClinVar annotation to yield the final data set of 46,935 pathogenic variants. b Pie charts visualizing the distribution of variant classes among all variants (left) and only among pathogenic variants identified in autosomal recessive disease genes (right). c The vast majority of pathogenic variants were very rare with global frequencies <0.001%, whereas only 82 and 5 pathogenic variants had frequencies between 0.1–1% and ≥1%, respectively. d Column plot showing the ten autosomal recessive disease associated genes with the highest and lowest number of pathogenic variants. Note that inter-gene differences in variant numbers exceed 500-fold. e The ten genes with the highest aggregated frequency of pathogenic variants are shown.