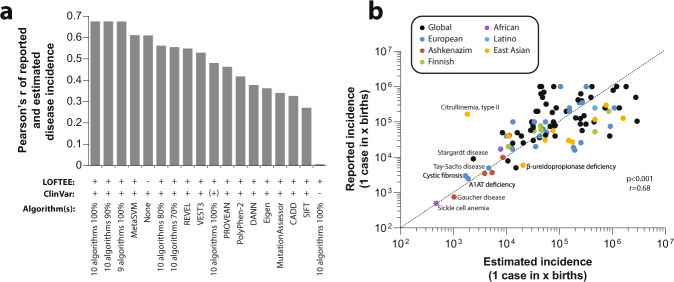
Fig. 2. Disease prevalence predicted on population-scale genomic data closely corresponds to clinically reported disease frequencies.
a Comparison of predictive performance of different prediction methods for 85 AR diseases. Note that integration of database information with stringent missense and nonsense predictions was overall most accurate, resulting in highly correlated predicted and reported disease incidence (Pearson’s r = 0.68; p < 0.0001). Note that when ClinVar annotations are not considered and disease incidence is purely calculated on the basis of computational pathogenicity assessments, the performance of the respective prediction model is very poor (r < 0.01). When only those pathogenic ClinVar variants are considered that are also estimated to be pathogenic using ten algorithms (indicated by “ClinVar (+)”), performance is strongly reduced (r = 0.48). b Detailed correlation of estimated and reported population-specific or global disease prevalence using the best performing prediction model from a ClinVar + ten algorithms + LOFTEE.