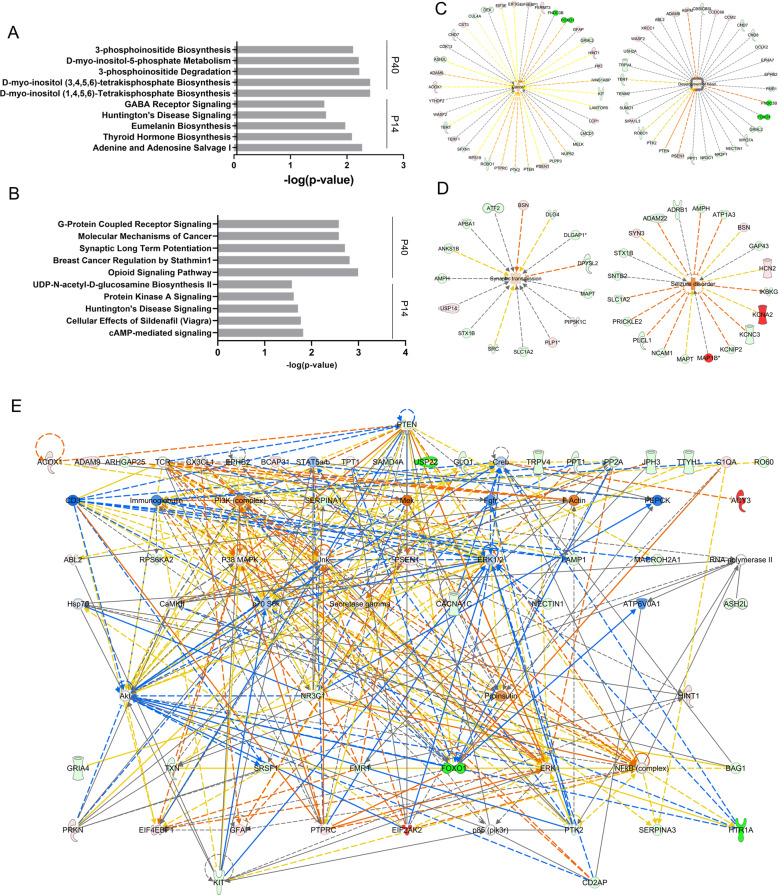
Fig. 3. Evaluation of the biological impact of the proteomic and phosphoproteomic changes in the Ptenm3m4/m3m4 brain.
A The top five most significant “Canonical Pathways” identified by IPA core analysis in the P40 cortical and P14 hemibrain proteome comparing Ptenm3m4/m3m4 mice to wildtype controls. B The top five most significant “Canonical Pathways” identified by IPA core analysis in the P40 cortical and P14 hemibrain phosphoproteome comparing Ptenm3m4/m3m4 mice to wildtype controls. C The top two “disease and bio function” networks (by size) identified by IPA core analysis from the P40 cortical proteome of Ptenm3m4/m3m4 mice compared to wildtype controls with activity prediction overlaid. Green = decreased expression. Red = increased expression. Color intensity = degree of increased/decreased expression. Hashed line = indirect biological relationship. Arrow = Direction of relationship. Orange = predicted activation. Yellow = observation inconsistent with direction of prediction. Gray = no influence on prediction. D The top two “disease and bio function” networks (by size) identified by IPA core analysis from the P40 cortical phosphoproteome of Ptenm3m4/m3m4 mice compared to wildtype controls with activity prediction overlaid. Green = decreased phosphorylation. Red = increased phosphorylation. Color intensity = degree of increased/decreased phosphorylation. Hashed line = indirect biological relationship. Arrow = Direction of relationship. Orange = predicted activation. Yellow = observation inconsistent with direction of prediction. Gray = no influence on prediction. E The top network identified by IPA from among the differentially expressed proteins with Molecular Activity Predictor (MAP) results overlaid. The network is organized hierarchically, meaning the most important regulatory molecule is placed on top. Green = decreased expression. Red = increased expression. Color intensity = degree of increased/decreased expression. Solid line = direct biological relationship. Hashed line = indirect biological relationship. Arrow = direction of relationship. Orange = predicted activation. Blue = predicted inactivation. Yellow = observation inconsistent with the direction of prediction. Gray = no influence on prediction.