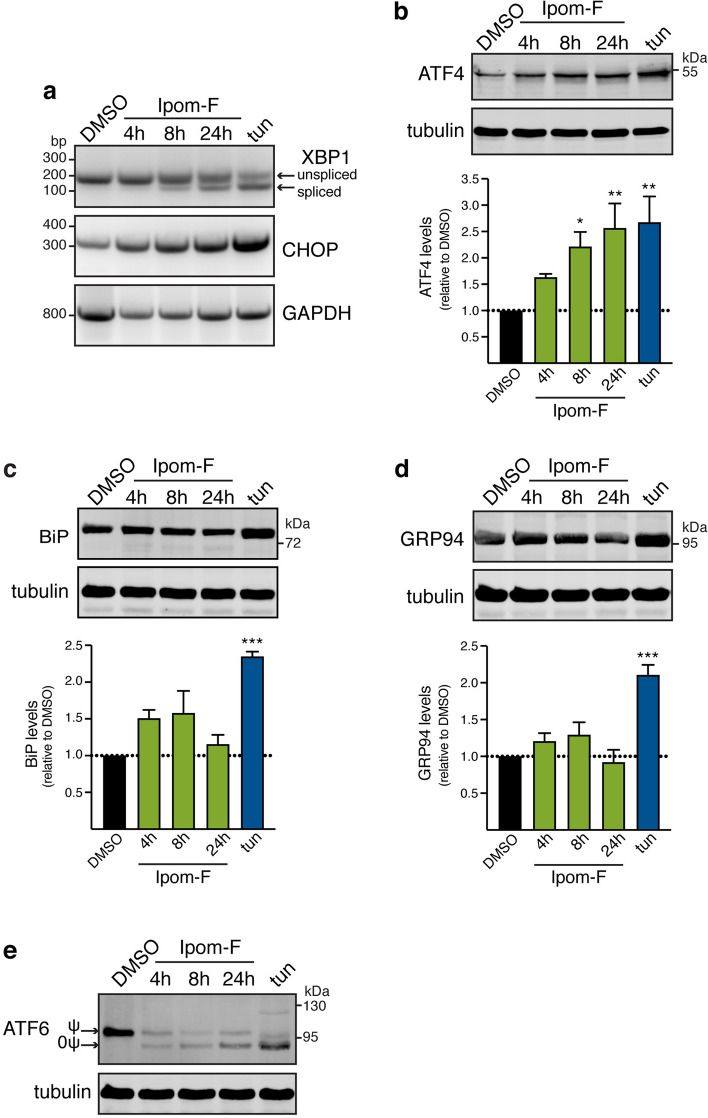
Figure 6.
Ipom-F activates an unconventional ER stress response. HepG2 cells were treated with DMSO for 24 h, tunicamycin (tun; 5 μg/ml) for 12 h or Ipom-F (100 nM) for the indicated times. (a) Total RNA was isolated from cells, and XBP1 mRNA splicing or levels of CHOP mRNA was determined by reverse transcription-PCR. Unspliced and spliced XBP1 mRNA are indicated. GAPDH served as a cDNA loading control. Full-length agarose gels are presented in Supplementary Fig. S7. (b–d) Detergent-soluble lysates were blotted for the indicated proteins. Infrared signals were quantified, normalised against the corresponding tubulin signals and expressed relative to the signal in DMSO-treated cells. Values are mean ± s.e.m; n = 3–4; *P < 0.05; **P < 0.01; ***P < 0.001 relative to DMSO (one-way ANOVA). Representative immunoblots are shown (full-length blots presented in Supplementary Fig. S6). (e) Total extracts were blotted for ATF6. Non-glycosylated (0ψ) and N-glycosylated (ψ) ATF6 are indicated. Please note that the immunoblot shown was probed with both rabbit anti-ATF6 and rat anti-Hsc70 antibodies (see Fig. 4c) using two-colour detection with an Odyssey Infrared Imager. Hence, the accompanying tubulin loading control is identical to that shown in Fig. 4c (full-length blots presented in Supplementary Fig. S6).