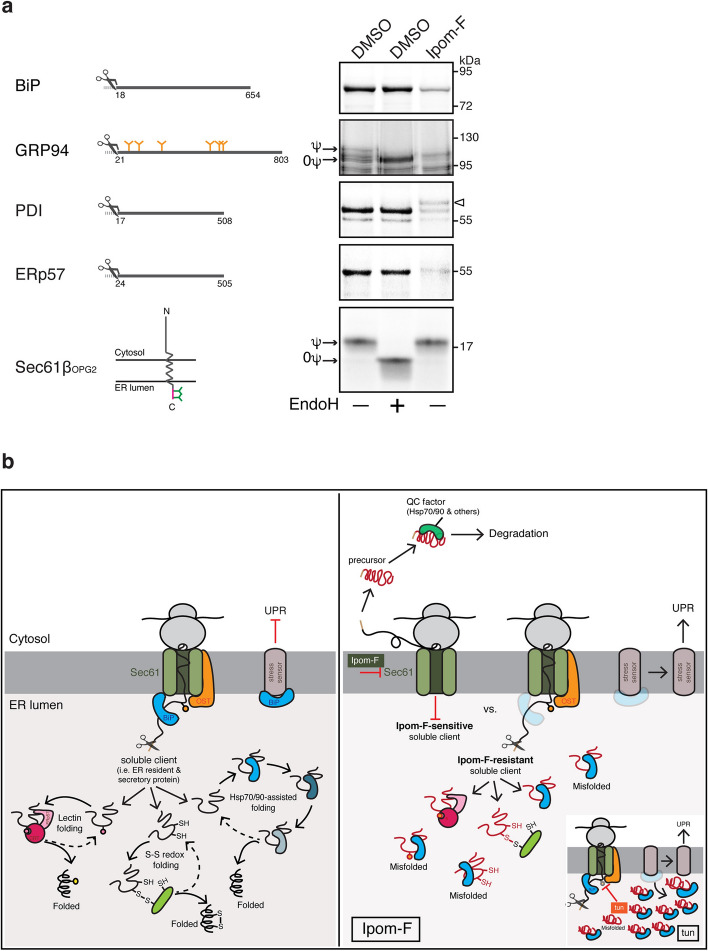
Figure 7.
Ipom-F compromises the translocation of soluble ER-resident chaperones and folding factors. (a) Model soluble ER-resident folding factors and the tail-anchored protein Sec61βOPG2 were translated in rabbit reticulocyte lysate supplemented with [35S]Met/Cys, canine rough microsomes and either DMSO or Ipom-F (1 μM), and the membrane-associated products were resolved by SDS-PAGE and analysed directly by phosphorimaging. Control samples were treated with EndoH to distinguish N-glycosylated (ψ) from non-glycosylated (0ψ) products. The position of the SP-uncleaved precursor of PDI is indicated with an arrowhead. Diagrams of preproteins showing the cleavage site of SPs (scissors symbol) and N-glycosylation sites (orange Y sites) are shown on the left. Representative phosphorimaging exposures are shown (full-length gels presented in Supplementary Fig. S5). (b) Proposed model of how the disruption of ER import by Ipom-F leads to ER stress. (Left) Under normal conditions, soluble ER clients, i.e. ER lumen residents and secretory proteins, are co-translationally translocated into the ER through the Sec61 translocon, often with the help from auxiliary components such as BiP. Shortly after, they engage ER lumen-resident chaperones and folding factors that promote protein folding and disulphide bond formation. ER stress sensors are maintained in an inactive form through BiP binding73,74. Protein N-glycosylation also occurs in the ER lumen during this phase of protein synthesis (not shown for simplicity). (Right) Ipom-F inhibits co-translational import of selected secretory proteins, such as α1AT, leading to the production of non-translocated precursors that mislocalise to the cytosol. These species appear to trigger the induction of cytosolic Hsp70/90 chaperones, which may contribute to a cytosolic quality control mechanism(s). Ipom-F-sensitive substrates also include central ER lumen-resident chaperones (BiP and GRP94) and other folding factors (PDI and ERp57), together with the type II integral membrane proteins ATF6, which acts as an ER stress transducer. These combined defects in ER translocation and ATF6-dependent stress signalling prevent the enhanced level of ER lumenal BiP that is seen after a tunicamycin-induced UPR and perturbs the folding of proteins such as FGA that still access the ER lumen in the presence of Ipom-F. These defects in ER function result in UPR signalling as evidenced by XBP1 mRNA splicing and increased levels of CHOP mRNA and ATF4 protein. (Right-inset) Tunicamycin inhibits protein N-glycosylation, which leads to the accumulation of incorrectly folded proteins in the ER. Increased demand for protein folding and degradation induces ER stress and activates UPR signalling. Dissociation of BiP from the ER stress sensors has been proposed to activate the UPR73,74.