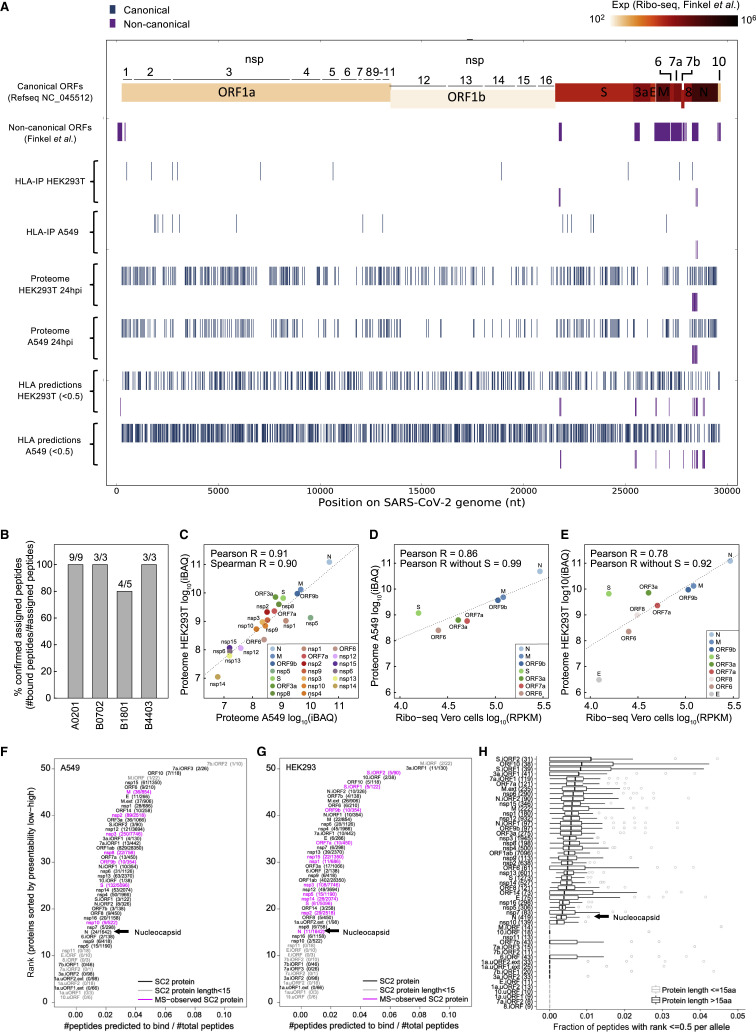
Figure 2.
SARS-CoV-2 HLA-I immunopeptidome and whole proteome
(A) Summary of peptide location across the SARS-CoV-2 genome from the HLA-I immunopeptidome, whole proteome, and predictions.
(B) Biochemical binding of HLA-I peptides to purified major histocompatibility complexes (MHCs). Shown are the fractions of peptides that were confirmed to bind the assigned alleles (half maximal inhibitory concentration [IC50] < 500 nM; Table S2).
(C) SARS-CoV-2 protein abundance in A549 and HEK293T cells 24 hpi. iBAQ, intensity-based absolute quantification.
(D and E) Comparison of our protein abundance measurements 24 hpi and Ribo-seq (Finkel et al., 2020b) in A549 (D) and HEK293T (E) cells.
(F) HLA-I presentation potential of SARS-CoV-2 ORFs in A549 cells. ORFs were ranked according to the ratio between the number of peptides predicted to bind any of the six HLA-I alleles in A549 and the total number of 8- to 11-mers.
(G) Similar to (F) for HEK293T cells.
(H) Presentation potential across 92 HLA-I alleles, shown as boxplots (median ratio, whiskers reach to lowest and highest values no further than 1.5× interquartile range [IQR] of the ratio between the number of peptides predicted to bind each allele and total number of peptides). SARS-CoV-2 ORFs are ranked by the median across HLA-I alleles.