Figure 6.
T cell responses to SARS-CoV-2 HLA-I peptides
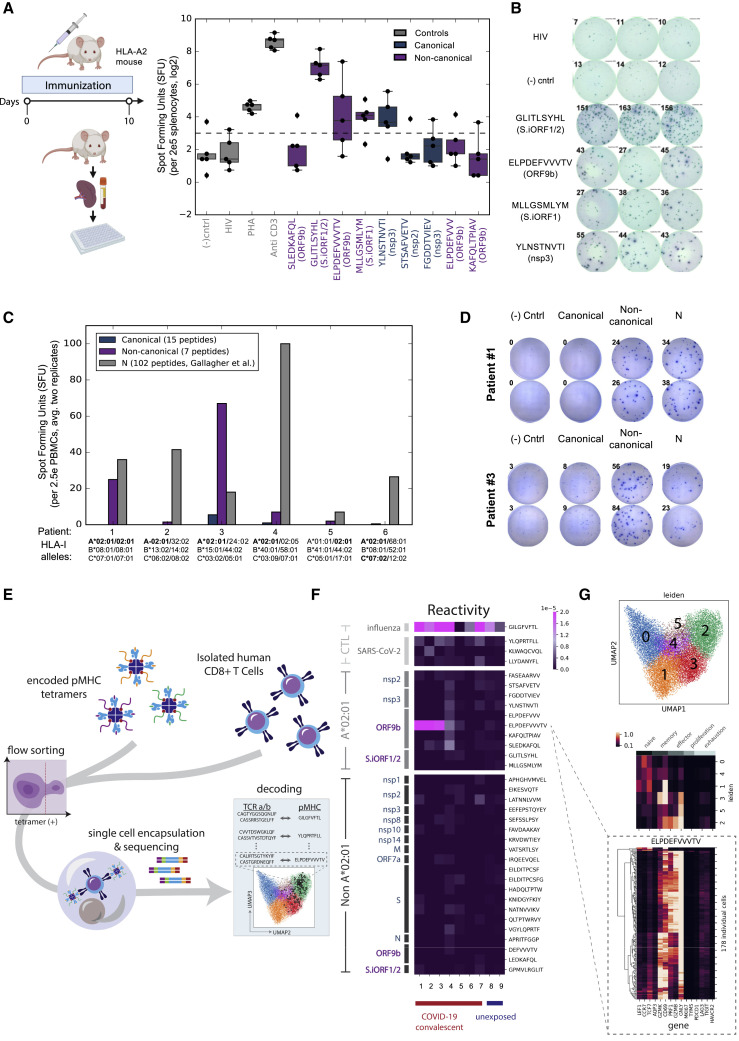
(A) Five HLA-A2 transgenic mice were immunized with a pool of nine HLA-I peptides detected on A∗02:01 in HEK293T cells for 10 days. Splenocytes were incubated with individual peptides and monitored for IFNγ secretion. HLA-A∗02:01 restricted HIV-Gag peptide and non-stimulated wells were used as negative controls. Anti-CD3 and phytohemagglutinin (PHA) were used as positive controls. The dashed line represents the threshold for positive responses (3× the median of the HIV-Gag). The box shows the quartiles, the bar indicates median, and the whiskers show the distribution.
(B) ELISpot images from one of the five vaccinated mice. Numbers indicate the spot count.
(C) PBMCs from convalescent COVID-19 individuals expressing A∗02:01 alleles were incubated with a pool of HLA-I peptides from canonical or out-of-frame ORFs. A pool of 102 peptides tiling the entire nucleocapsid (N) protein that was evaluated in the same samples (Gallagher et al., 2021) served as positive control. Bars show the mean of duplicates.
(D) ELISpot images of individuals #1 and #3. Numbers indicate the spot count.
(E) Illustration of the multiplexed tetramer assay and T cell single-cell profiling.
(F) CD8+ T cell reactivity detected in convalescent COVID-19 individuals and unexposed subjects expressing A∗02:01 to individual HLA-I peptides. The score in the heatmap indicates the fraction of peptide-specific reacting T cells from total CD8+ cells in the sample.
(G) Single-cell transcriptomics of reactive T cells. Top panel: uniform manifold approximation and projection (UMAP) embedding of all tetramer-positive cells colored by unsupervised clustering. Center panel: expression levels of 15 genes associated with different states of T cells, as characterized previously (Su et al., 2020). Bottom panel: expression level of these 15 genes in individual T cells reactive to ELPDEFVVVTV peptide from ORF9b.