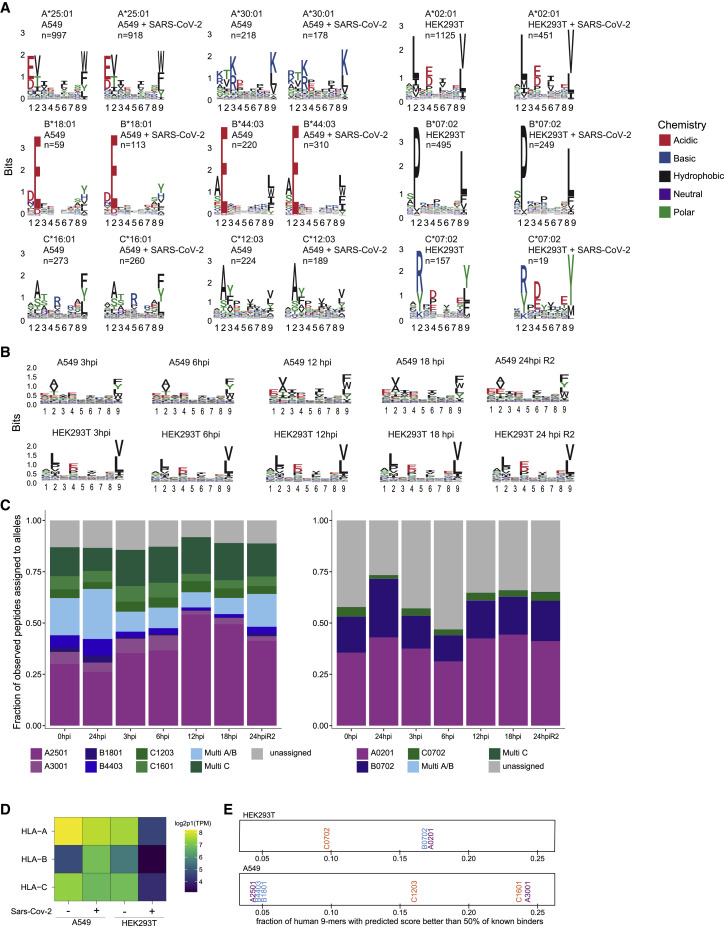
Figure S2.
Peptide logos and allele assignment for all experiments, related to Figure 1
(A) Logo plots for individual alleles of peptides identified and assigned to cell line specific alleles with HLAthena percentile rank < 0.5 for naive and 24h post Sars-CoV-2 infected A549 (left) and HEK293 (right) cells. (B) Peptide logo plots aggregated over all alleles for label free time course experiments in A549 and HEK293 samples. (C) Allele assignment for peptides identified in time course experiments using HLAthena with a percentile rank < 0.5 cutoff. (D) Expression level of HLA-A, -B, and -C alleles as measured by RNA-seq in A549 and HEK293T cell lines pre- and 24hr post-infection. (E) All 9-mer peptides tiled along human protein sequences were predicted for binding to the HLA alleles present in HEK293T (top) and A549 (bottom) cell lines. Per allele, the fraction of those 9-mers with predicted binding scores that are better than 50% of previously identified known binders in mono-allelic experiments for the allele is shown.