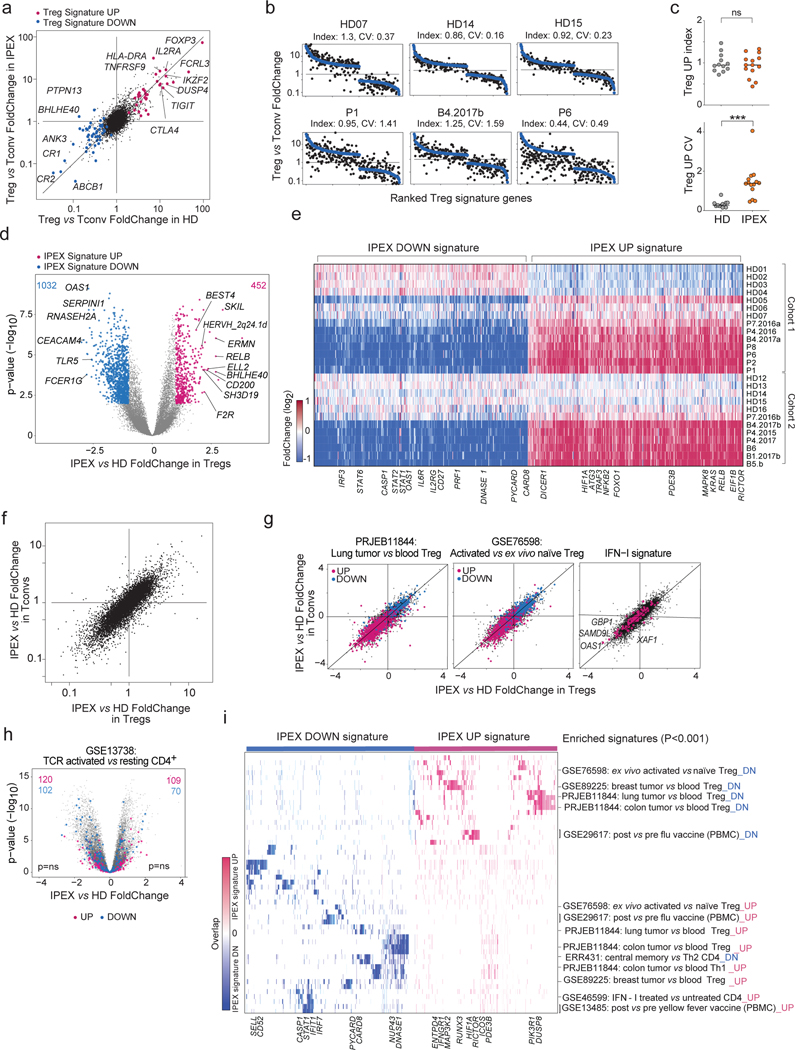
Fig. 2. Transcriptional changes in IPEX Treg and Tconv cells by population RNAseq.
Population RNAseq was performed on sorted Tconv and Treg-like cells from HD (n=12) and IPEX (n = 10) donors. a. The Treg/Tconv FoldChange in HD (x-axis) and IPEX (y-axis); Treg signature 5 genes are highlighted.
b. Ranked FC plots of Treg signature transcripts for individual donors, ranked according to mean FC in all HD (blue dots). FC values for each donor (black dots) computed from the donor’s Treg vs the mean of HD Tconv.
c. Index and coefficient of variation (CV) of Treg Up signature transcripts (each dot is a sample). *** two-sided t.test p < 0.001.
d. FoldChange vs p value (volcano) plot comparing normalized expression in all IPEX to all HD samples. Genes with differential expression (two-sided t.test p<0.01, FC >2) are highlighted (452 up genes and 1032 down genes), and numbers shown.
e. Heatmap of the expression ratio, for IPEX signature genes defined in d, in Tregs from each donor, computed against the mean expression in HD Tregs (each cohort computed against its own HD Treg set).
f. Comparison of mean IPEX effect (all IPEX vs all HD samples) in Treg (x-axis) versus Tconv (y-axis).
g. Same FC/FC plot as in f, but highlighted with representative signatures of tumor-infitrating Tregs, activated Treg vs resting Treg, and IFN-I induced genes.
h. Same volcano plot as in d, but with highlights from a representative CD4+ T activation signature (ns, hypergeometric test).
i. Heatmap, for the IPEX signature genes defined in d, of their overlap with the pathways and signatures that significantly overlap with the IPEX signature (hypergeometric test p<0.001). Present genes are shown by tick marks, color-coded by their IPEX/HD FoldChange.