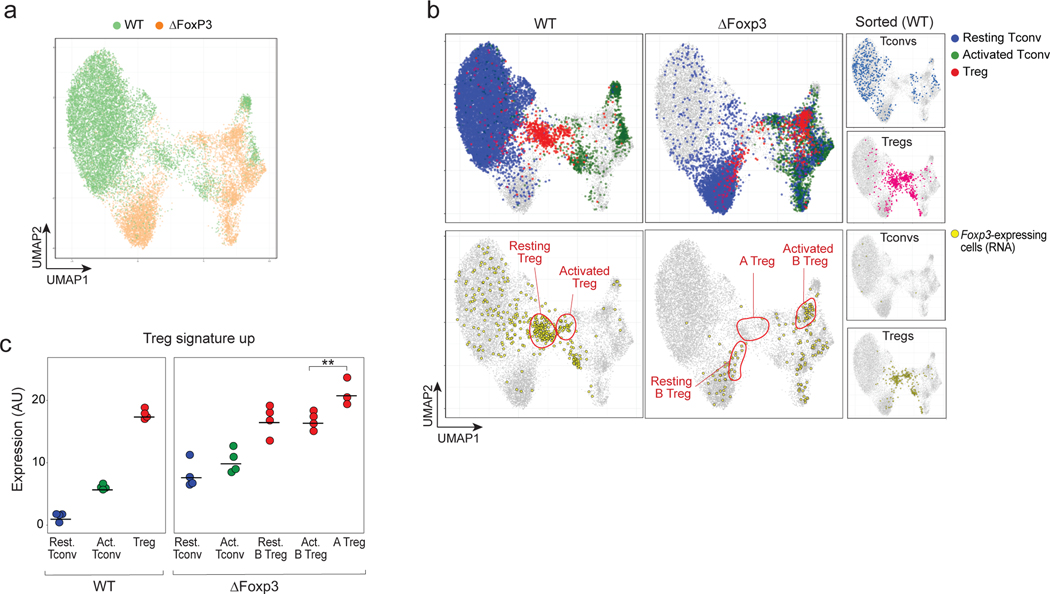
Fig. 5. scRNAseq reveals heterogeneous effects of the Foxp3 ablation in ΔFoxp3 mice.
scRNAseq was performed on CD4+ T cells from WT (n = 4) and ΔFoxp3 (n = 4) mice (18,097 cells altogether).
a. 2D UMAP plot of CD4+ single-cell transcriptomes for WT (green) and ΔFoxp3 (orange)
b. Same UMAP as b. Top row: resting Tconv (blue), activated Tconvs (green), and Tregs (red) (assigned by graph-based clustering on the merged dataset after regressing out the ΔFoxp3 signature, see Methods) are highlighted; bottom: cells with an active Foxp3 locus (GFP or Cre transcripts detected) are colored yellow. The small inserts on the right show sorted control WT Tconvs and WT Tregs, sorted and included as spike-in controls.
c. Treg signature expression in resting Tconvs, activated Tconvs, and Tregs in WT and ΔFoxp3 mice (normalized counts). ** p < 10–2, two-sided t.test.