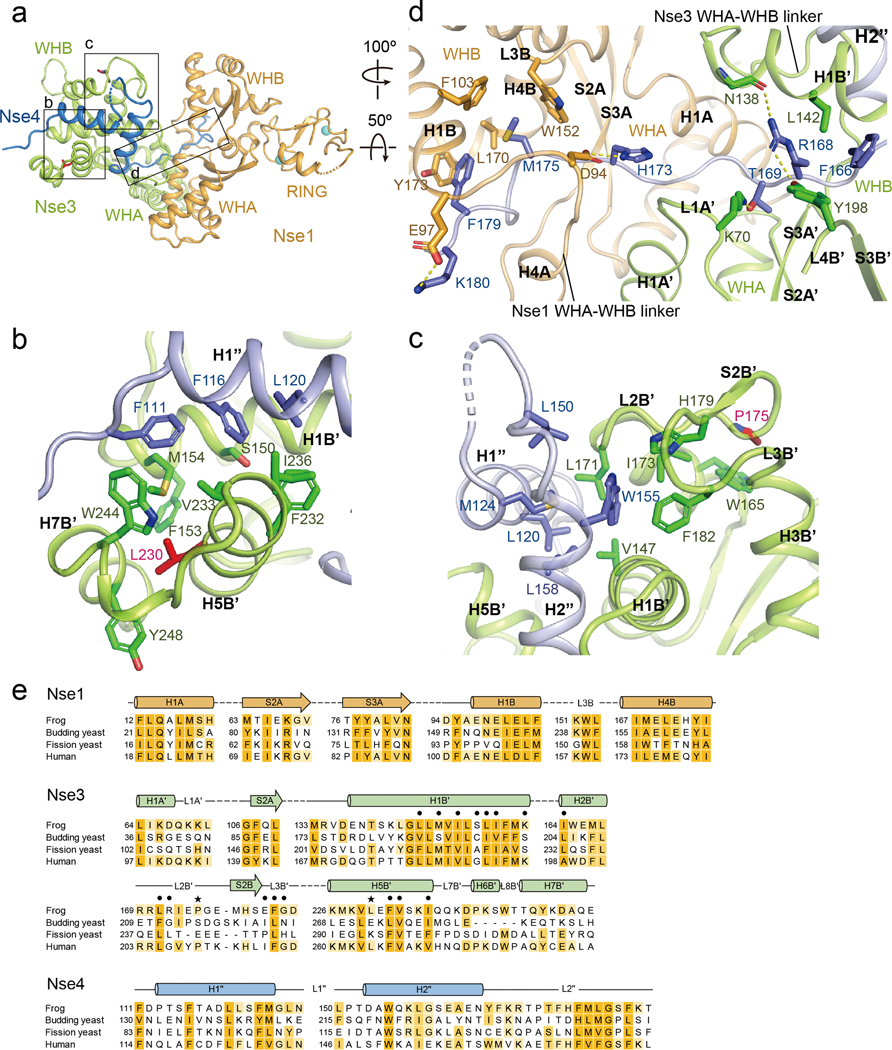
Fig. 3.
The three interfaces between XlNse4 and the XlNse1-Nse3 dimer.
(a) Overall structure of the Nse1 (orange)-3 (green)-4 (blue) complex. Three interfaces are highlighted with boxes.
(b) The first interface, between the first helix of Nse4 (blue) and the WHB of Nse3 (green). Residues in the H1” helix of Nse4 are surrounded by hydrophobic residues in the H1B’, H5B’, and H7B’ helices of Nse3. Leu230, a mutated residue in LICS is shown in a red stick. The interacting residues in Nse3 and Nse4 are also marked in the sequence alignment in Fig. S3 and S4.
(c) The second interface, between the second helix of Nse4 (blue) and the H1B’ and H5B’ helices, S2B’ strand, and L2B’ and L3B’ loops of Nse3 (green). Pro175, a mutated residue in LICS is shown in a red stick. See also Fig. S3 and S4.
(d) The third interface, between Nse4 (the extended loop) and the Nse1-Nse3 complex. The view is 100º and 50º-rotated along the y and the x axis, respectively, relative to that in (a). The L2” loop of Nse4 (blue) interacts with Nse1 (orange) and Nse3 (green) by threading through their interface. The first half of the Nse4 L2” loop is surrounded by Nse3, as well as Nse1 (right). The second half of Nse4 L2” loop is surrounded primarily by Nse1 (left).
(e) Conservation of the residues in the Nse1-3-4 complex in (b)-(d). The point mutations sensitive to DNA damage inducing agents in S. pombe are indicated by dots. LICS related mutations are marked by black stars.