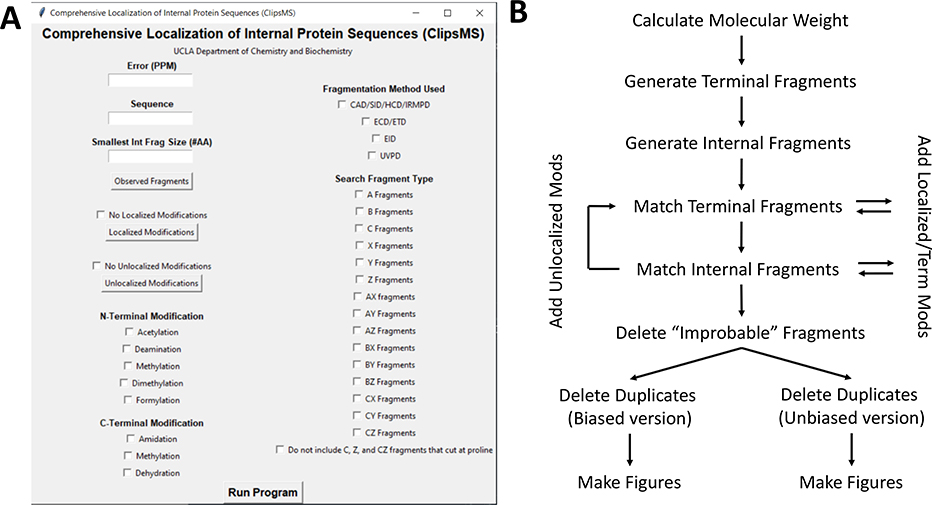
Figure 1.
A. The graphical user interface (GUI) for ClipsMS. The user can input several key parameters including the error allowed, the smallest internal fragment size, the sequence, the observed fragments, any modifications on the sequence and the type of fragments to search. B. The workflow of the algorithm and how it matches peaks input by the user. The algorithm calculates all theoretical terminal and internal fragments, matches all peaks, makes decisions on which assignments to keep, and automatically generates figures.