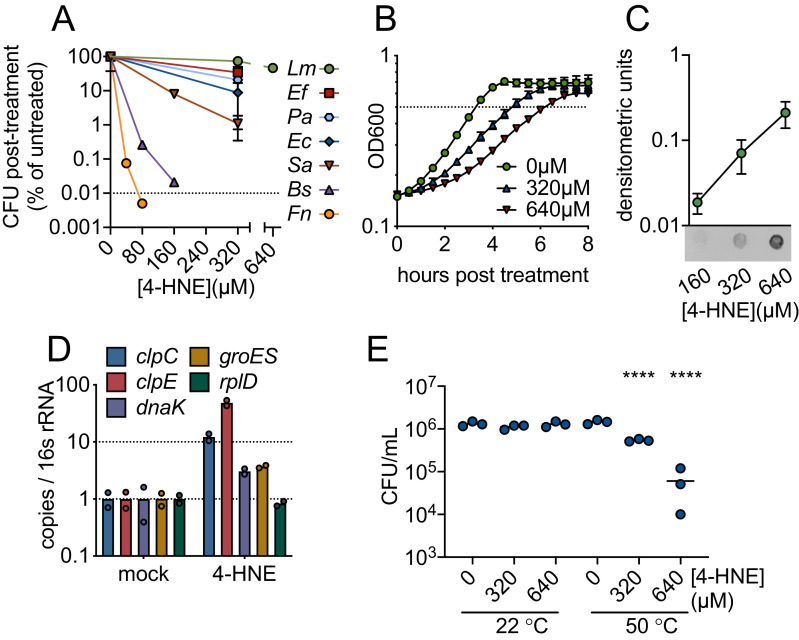
Figure 2. 4-HNE is a bactericidal, cell-permeable and protein damaging molecule.
(A) Survival of mid-log (0.4–0.8 OD600) Listeria monocytogenes (Lm), Enterococcus faecalis (Ef), Pseudomonas aeruginosa (Pa), Escherichia coli (Ec), Staphylococcus aureus (Sa), Bacillus subtilis (Bs), and Francisella novicida (Fn) following exposure to various concentrations of 4-HNE or mock vehicle (ethanol) in PBS at 37°C for 1 hr. Data are reported as recovered CFU normalized to mock-treated controls. Dashed line is at the limit of detection. (B) Growth of L. monocytogenes in TSB at 37°C with various concentrations of 4-HNE added at time zero. Dashed line at OD600 0.5 (C) Anti-4-HNE dot blot of soluble bacterial lysates from mid-log L. monocytogenes resuspended in PBS and treated with increasing concentrations of 4-HNE for 30 min. Protein levels were normalized (3 µg total protein) and signal quantified by densitometry on a Licor Odyssey Fc. (D) RT-qPCR measurement of expression of the indicated genes from mid-log L. monocytogenes in TSB treated with 640 µM 4-HNE for 20 min. Expression normalized to 16S rRNA levels. (E) Recovered CFU of WT L. monocytogenes following exposure to 4-HNE or mock vehicle (ethanol) in PBS at 37°C for 1 hr, followed by heat shock (50°C) or no heat shock (22°C) treatment for 10 min. Data in figures (A) and (B) are in biological triplicate. Data in (C) and (D) are biological duplicate. Data in (E) are in technical triplicate and representative of at least two independent experiments. Statistics in (E) are an ordinary one-way ANOVA with a Dunnett’s multiple comparison test against each untreated. Error bars are mean ± SD. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. In figure (E), the line is drawn at the median of data.