Figure 1 |. A library of cancer-associated histone mutations.
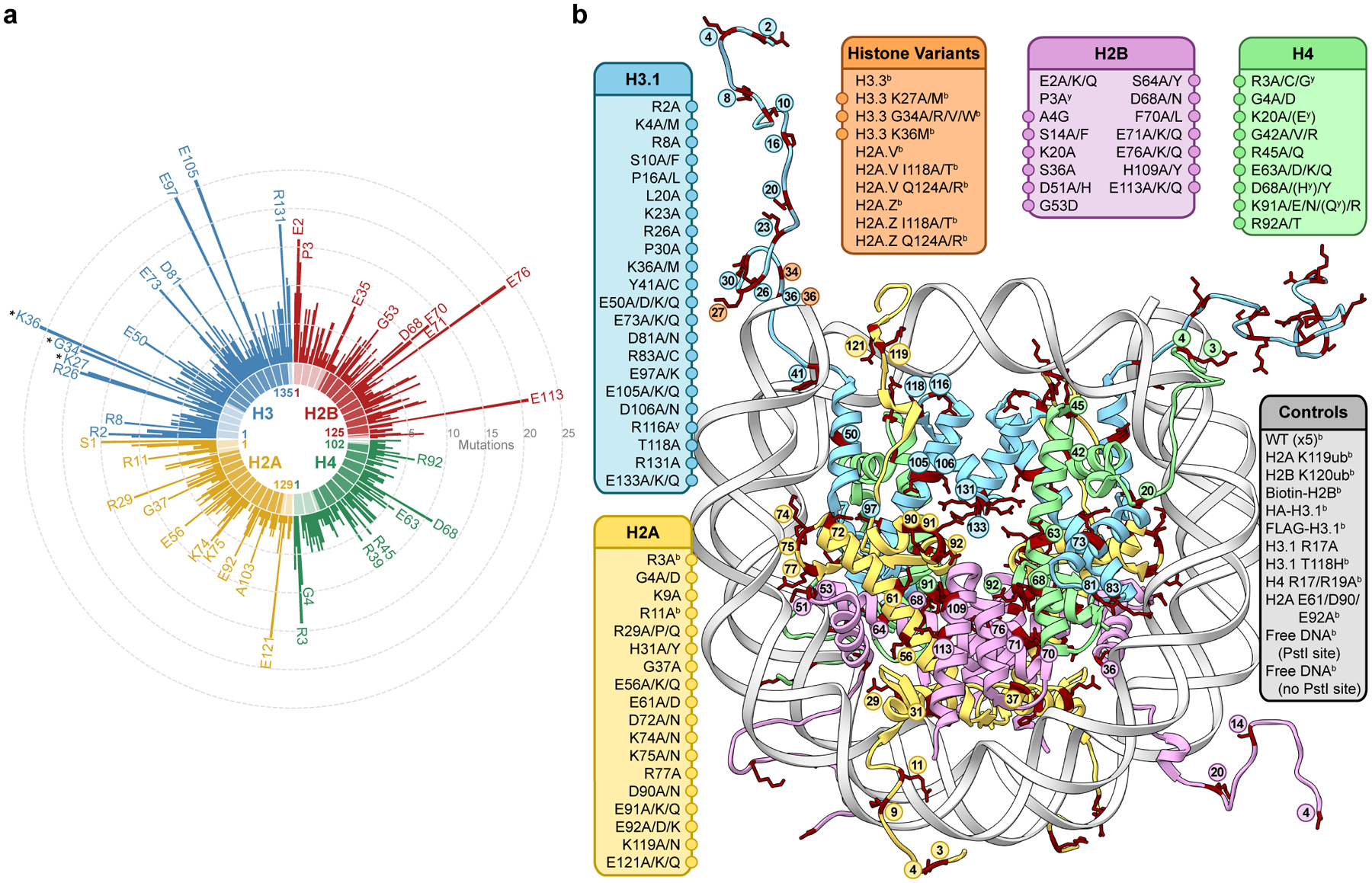
a, Circular barplot showing mutation counts in the four core histones in human cancers (data from Nacev et al., 2019)19. Select highly mutated sites are labeled, and established oncohistone mutations are denoted with an asterisk. Darker and lighter regions in each histone represent the globular and tail domains, respectively. b, Diagram depicting all histone mutants, variants, and controls included in the barcoded mononucleosome and humanized yeast libraries. Mutations with adjacent circles are displayed on the nucleosome structure with coordinated colors and labeled by residue. In addition to any specific missense mutations that were overrepresented in the dataset, alanine mutations were included at every site, offering both experimental redundancy as well as the ability to distinguish between loss and gain of function at a given amino acid residue. Indicated mutants are found only in the barcoded mononucleosome (b) or humanized yeast libraries (y), respectively (PDB 1kx5).
