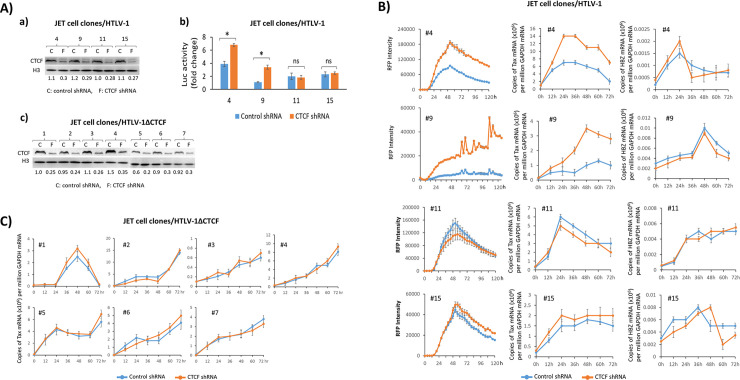
Fig 5. Inhibition of CTCF binding to HTLV-1 provirus affects proviral gene transcription.
Clonal HTLV-1 (A-a and B) or HTLV-1ΔCTCF (A-c and C) infected JET cell lines were generated, each carrying a single, latent provirus at a unique integration site. The infected JET cells from panels B, and C were collected and cellular CTCF was detected by Western blotting using anti-CTCF antibody. Histone H3 was also blotted as a loading control. The amount of CTCF was measured by densitometry quantification using BioRad image lab software, normalized to the value of histone H3 and is indicated under each lane. A-b). The JET cells from Fig 5B were cocultured with Jurkat cells expressing firefly luciferase driven by the HTLV-1 LTR (LTR-Luc) in the presence or absence of PMA/Ionomycin for 48 hr and viral infectivity was determined by measuring luciferase activity and presented as a fold change compared with the values of untreated cells (t test: *p < 0.05; ns: not significant). B & C). Viral gene expression was activated with PMA/Ionomycin and monitored by measuring Tax mediated RFP production using the IncuCyte live cell image system. The levels of tax and hbz mRNA were also measured by qRT-PCR. Total copy number of each mRNA was determined using plasmid DNA standards and normalized to 106 copies of GAPDH mRNA. Cell clone number of HTLV-1 infected and HTLV-1ΔCTCF cells are indicated in each panel. Each experiment was repeated three times and a representative result with three replicates is shown with standard errors.