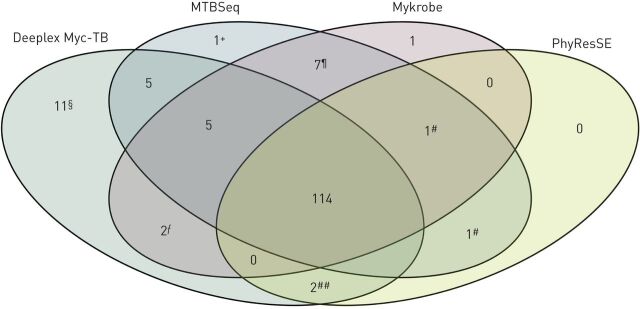
FIGURE 3.
Venn diagram representing the agreement between resistant phenotypes identified by four Mycobacterium tuberculosis resistance and susceptibility prediction tools: Deeplex Myc-TB, MTBSeq, Mykrobe and PhyResSE. WGS: whole-genome sequencing. The numbers of resistant phenotypes predicted by Deeplex Myc-TB analysis on 109 sputum samples from Djibouti and other analysis tools fed with WGS data from corresponding cultures are shown. #: two rifampicin resistance phenotypes predicted by MTBSeq and PhyResSE and/or Mykrobe based on rpoB S431T and D435V, reflecting probable WGS or culture contaminations (see text); ¶: seven pyrazinamide resistance phenotypes predicted for “Mycobacterium canettii”-containing cultures by MTBSeq and Mykrobe based on panD M117T and pncA A46A, respectively; +: one pyrazinamide resistance phenotype predicted by MTBSeq based on pncA D136G; §: 11 resistant phenotypes predicted by Deeplex Myc-TB based on 10 minority variants (3–12%) and one ethA frameshift-causing indel; ƒ: two streptomycin resistance phenotypes predicted by Deeplex Myc-TB and Mykrobe based on gidB G69D; ##: two ethambutol resistance phenotypes predicted by Deeplex Myc-TB and PhyResSE based on embB S297A and Y319S.