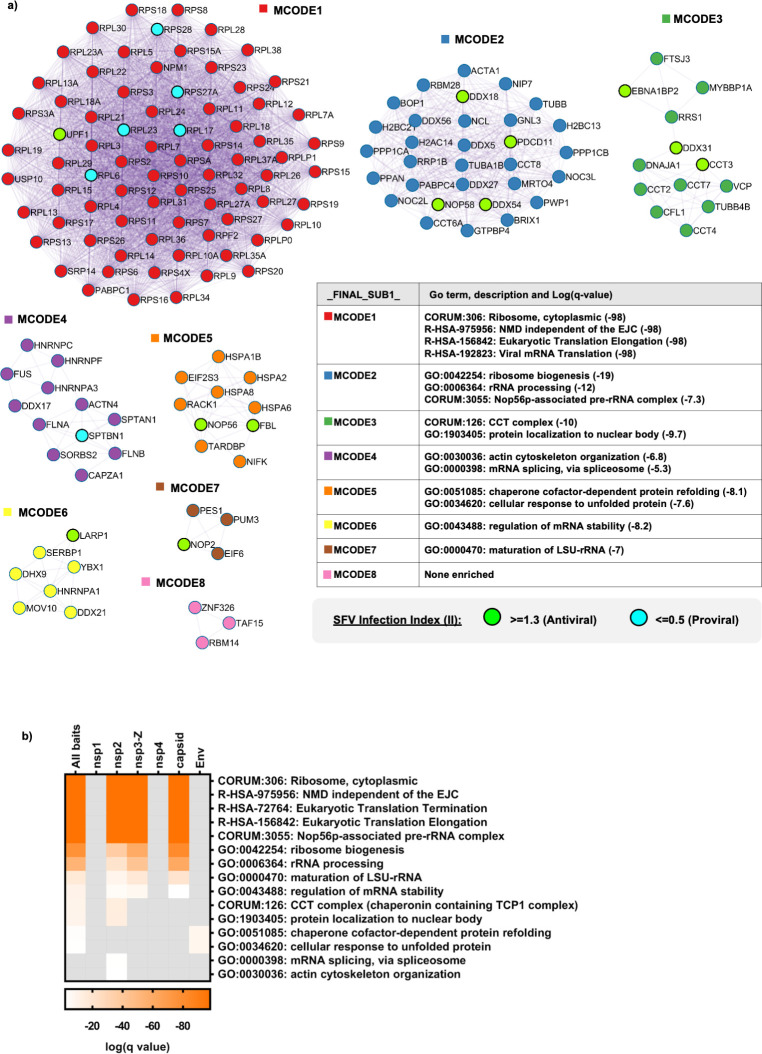
Fig 4. SFV interactors are highly enriched for GO terms related to translation, NMD and ribosome biogenesis.
a, Protein-protein interaction enrichment analysis for the full list of SFV interactors was carried out using Metascape with the following databases: BioGrid, InWeb_IM and OmniPath. Densely connected network components were identified by applying the Molecular Complex Detection (MCODE) algorithm and the resultant MCODE networks are displayed. In addition, the proteins identified as having an effect on SFV replication through the siRNA screen are shown within the MCODE complexes (green and turquoise circles). In order to assign biological meanings to the MCODE networks, GO enrichment analyses were performed using the following sources: KEGG Pathway, GO Biological Processes, Reactome Gene Sets, Canonical Pathways and CORUM. All genes in the genome were used as the enrichment background. Between 1–4 GO terms were chosen to represent each MCODE network, with the relevant significance values depicted alongside them. b, Heat map comparing the significance of GO terms (chosen in a) among the different SFV baits. GO enrichment analysis was applied to the set of MCODE networks identified for each individual list (nsp1, nsp2, nsp3-Z, nsp4, capsid and Env), as well as for the merged list (All baits) and the significance values depicted in the heat map.