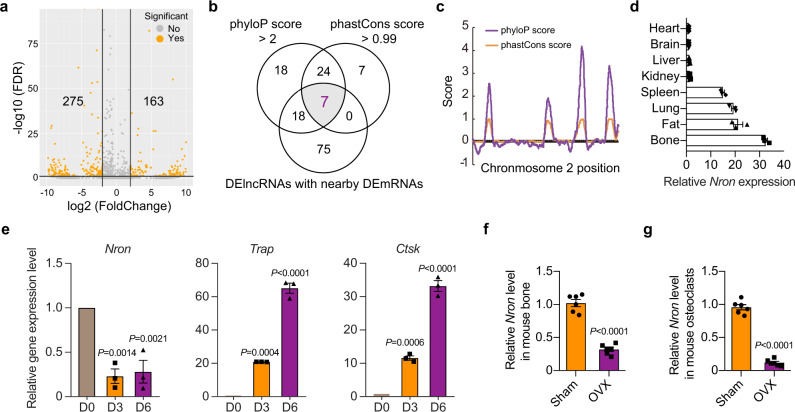
Fig. 1. Identification of Nron as a candidate regulatory lncRNA in osteoclasts.
a Volcano plots of the differential lncRNAs expression in undifferentiated BMMs and BMMs-derived osteoclasts. b Venn diagram representing the selection strategy for conserved lncRNAs. c PhastCons and PhyloP score of Nron. d Q-PCR analysis of Nron expression in eight types of C57BL/6 J mouse tissue, (n = 3/group). e Q-PCR analysis of Nron, Trap, and Ctsk gene expression levels during BMMs osteoclast differentiation, (n = 3 biologically independent experiments). f Q-PCR analysis of Nron expression in whole bone specimens (without bone marrow) from sham and ovariectomized (OVX) mice, (n = 6/group). g Q-PCR analysis of Nron expression in CTSK + osteoclasts isolated by microdissection from Sham and OVX mice, (n = 6/group). Significant differences between two groups were determined by unpaired Student’s t-test (two-tailed) and significant differences among multiple groups were determined by one-way ANOVA with Dunnett’s multiple comparisons test. Data are presented as the means ± s.e.m. Source data are provided as a Source Data file.