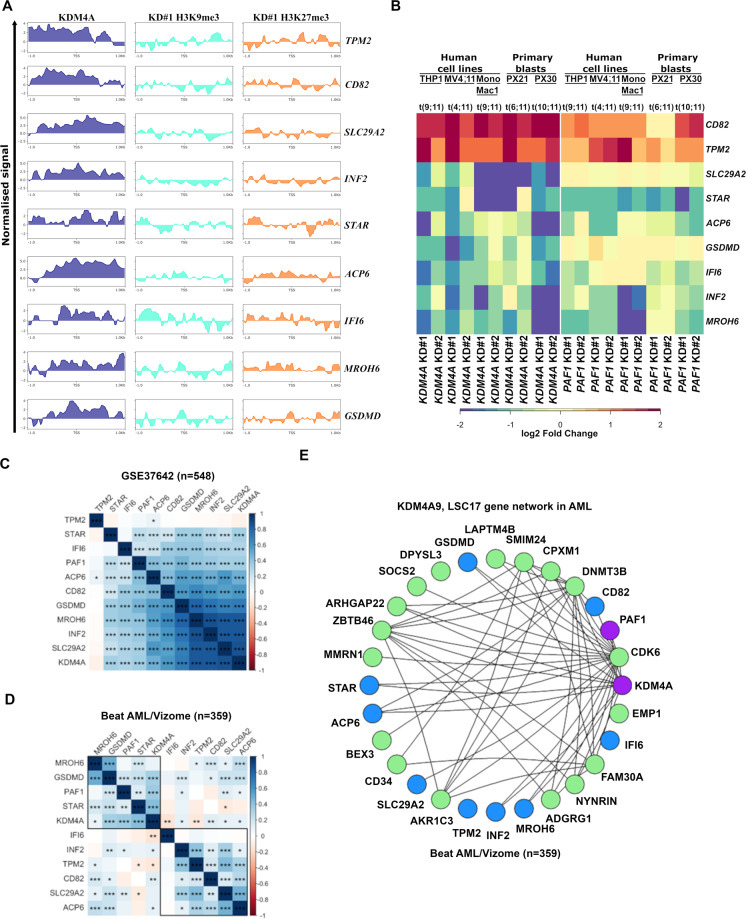
Fig. 8. KDM4A-mediated epigenomic network required for AML cell self-renewal and survival.
A Heatmap showing relative expression of KDM4A-9 signature genes as determined by QPCR in the indicated human MLLr-AML cell lines and AML primary cells following KDM4A KD or PAF1 KD in comparison with NTC control cells (n = 3). B Input normalized ChIP-seq coverage tracks showing KDM4A ChIP signal in WT THP1 cells and H3K9me3/H3K27me3 ChIP signal normalized to NTC in KDM4A KD THP1 cells at KDM4A-9 signature genomic loci (+/−1 kb TSS). The normalized signal shown is the log2 ratio of read counts compared against input control. C–D Correlation matrices showing the Pearson correlation coefficients for KDM4A, KDM4A-9 genes, and PAF1 gene expression in GSE37642 (C) and Beat AML/Vizome (D) AML datasets. Significance determined by t-test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. E KDM4A, PAF1, KDM4A-9 (in blue), and LSC17 (in green) gene network showing the topological overlap between genes as detected from 262 AML samples (Beat AML) (the corresponding topological overlap matrix (TOM) ≥ 0.05 between nodes. Genes with high topological overlap matrix (TOM) measure (TOM ≥ 0.05) have related functions).