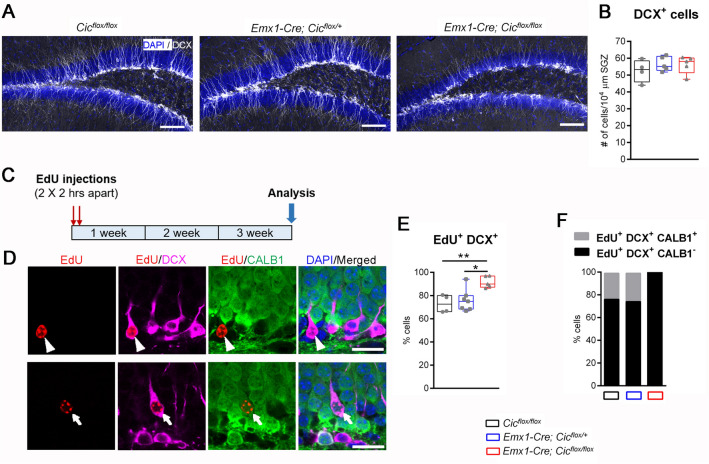
Figure 3.
Impaired development of DCX+ cells in the Emx1-Cre Cic knockout mice. (A) Representative images of the dentate gyrus in the three mouse lines show severe dendritic morphology defects in DCX+ cells of the knockout mice. DAPI (blue) and DCX (grey). Scale bars = 100 µm. (B) Quantification of the total number of DCX+ cells in the three genotypes. No statistically significant difference was found. (C) A schematic of the EdU-labelling study. Mice were injected with two doses of EdU that were separated by two hours and were analysed three weeks post injections. (D) Representative images of the subgranular zone three weeks post EdU injection showing staining for EdU (red), DCX (magenta), CALB1 (green), and DAPI (blue). The arrowhead points to an EdU-labelled cell that is positive for DCX but negative for CALB1. The arrow points to an EdU-labelled cell that is double positive for DCX and CALB1. Scale bars = 20 µm. (E) Quantification of the percentage of EdU-labelled cells that were DCX+ in mice of the three genotypes. (F) Quantification of the relative proportion of EdU+ DCX+ cells that were CALB1- or CALB1+ in mice of the three genotypes. P < 0.0001 using Chi-square test. N = 4–7 animals per group. In (B) and (E), data are presented in box-and-whisker plots showing all data points, where centre lines represent medians, box limits represent interquartile ranges, and whiskers represent minimum to maximum data ranges. Statistical analyses were performed with one-way ANOVA with Tukey’s post hoc test. *P < 0.05; **P < 0.01.