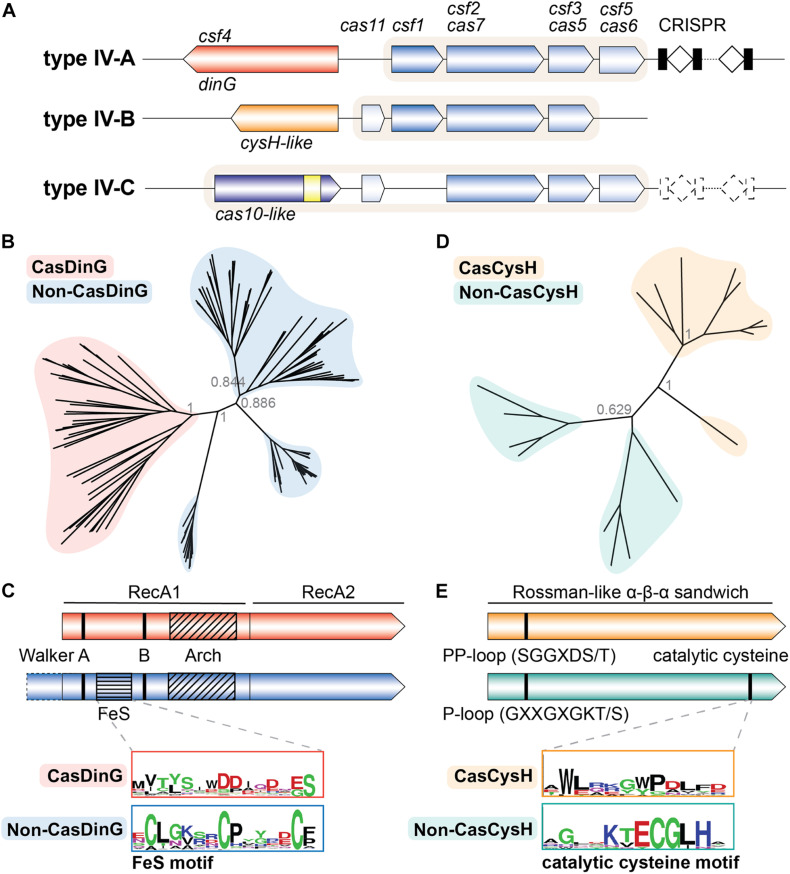
FIGURE 1.
The type IV Cas accessory proteins have evolved a Cas specific function. (A) Classification schematic of type IV CRISPR-Cas systems. A typical locus is represented for each type IV subtype. Dashed lines indicate components that are sometimes not encoded by the subtype. Shaded backgrounds highlight which gene products form the ribonucleoprotein (RNP) complex. The yellow square in the IV-C cas10-like large subunit represents an HD nuclease domain. (B) Phylogenetic tree of Cas- and non-CasDinG sequences. Posterior probabilities are shown. (C) Cartoons of Cas- and non-CasDinG sequences indicating positions of certain helicase motifs and domain architecture. Weblogos (Crooks, 2004) of the FeS cluster region in non-CasDinG (below, blue outline) and CasDinG (top, red outline) are shown. (D) Phylogenetic tree of Cas- and non-CasCysH sequences. Posterior probabilities are shown. (E) Cartoons of Cas- and non-CasCysH sequences. CasCysH is predicted to adopt the Rossman-like α - β - α fold observed in non-CasCysH structures. Positions and sequences of P- and PP-loops are indicated. Weblogos of the catalytic cysteine in non-CasCysH (bottom, teal outline) and CasCysH (top, orange outline) are shown.