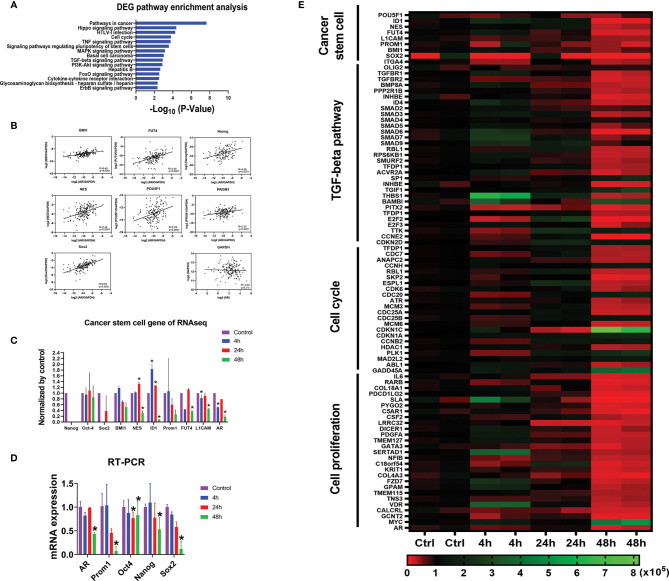
Figure 5.
AR positively correlates with CSC marker genes at mRNA expression level. (A) Gene Set Enrichment Analyses (GSEA) against the KEGG pathway database showing the top 15 pathways most significantly regulated in the U87MG cells with differentially expressed genes (DEGs) after the treatment of enzalutamide for 48 h. (B) The positive correlations of mRNA expressions between CSC marker genes and AR from in vivo RNA-seq results in GBM patients from TCGA database. GAPDH, a housekeeping gene, showed no correlation with AR (R = -0.03). (C) mRNA expression levels of various CSC marker genes in U87MG cells cultured in vitro before and after the treatment of enzalutamide (80 µM) from RNA-seq results from our laboratory. CD133 is an alias name for Prom1. (D) Quantitative RT-PCR results of the representative CSC genes after the treatment of enzalutamide. (E) Heatmap of the genes specific for cancer stem cell, TGF-β signaling pathway, cell cycle, and cell proliferation responding to enzalutamide treatment in U87MG cells at different time points. The scale bar at the bottom represents the normalized read counts of the genes from RNA-seq results. *p < 0.05.