Abstract
We have previously established that PV+ neurons and Npas1+ neurons are distinct neuron classes in the external globus pallidus (GPe): they have different topographical, electrophysiological, circuit, and functional properties. Aside from Foxp2+ neurons, which are a unique subclass within the Npas1+ class, we lack driver lines that effectively capture other GPe neuron subclasses. In this study, we examined the utility of Kcng4-Cre, Npr3-Cre, and Npy2r-Cre mouse lines (both males and females) for the delineation of GPe neuron subtypes. By using these novel driver lines, we have provided the most exhaustive investigation of electrophysiological studies of GPe neuron subtypes to date. Corroborating our prior studies, GPe neurons can be divided into two statistically distinct clusters that map onto PV+ and Npas1+ classes. By combining optogenetics and machine learning-based tracking, we showed that optogenetic perturbation of GPe neuron subtypes generated unique behavioral structures. Our findings further highlighted the dissociable roles of GPe neurons in regulating movement and anxiety-like behavior. We concluded that Npr3+ neurons and Kcng4+ neurons are distinct subclasses of Npas1+ neurons and PV+ neurons, respectively. Finally, by examining local collateral connectivity, we inferred the circuit mechanisms involved in the motor patterns observed with optogenetic perturbations. In summary, by identifying mouse lines that allow for manipulations of GPe neuron subtypes, we created new opportunities for interrogations of cellular and circuit substrates that can be important for motor function and dysfunction.
SIGNIFICANCE STATEMENT Within the basal ganglia, the external globus pallidus (GPe) has long been recognized for its involvement in motor control. However, we lacked an understanding of precisely how movement is controlled at the GPe level as a result of its cellular complexity. In this study, by using transgenic and cell-specific approaches, we showed that genetically-defined GPe neuron subtypes have distinct roles in regulating motor patterns. In addition, the in vivo contributions of these neuron subtypes are in part shaped by the local, inhibitory connections within the GPe. In sum, we have established the foundation for future investigations of motor function and disease pathophysiology.
Keywords: arkypallidal neurons, behavioral dynamics, body kinematics, local collaterals, machine learning, reciprocal inhibition
Introduction
The basal ganglia are a network of subcortical structures that are involved in motor control and adaptive behavior. Dysfunction within this circuit can be devastating, as seen in patients afflicted with Parkinson's disease (Albin et al., 1989; DeLong, 1990; Graybiel et al., 1994; Mink, 1996; DeLong and Wichmann, 2007; Redgrave et al., 2010; Nelson and Kreitzer, 2014; Jahanshahi et al., 2015; Dudman and Krakauer, 2016; Grillner and Robertson, 2016; Klaus et al., 2019; Park et al., 2020). The external globus pallidus (GPe) is reciprocally connected with the dorsal striatum and subthalamic nucleus (STN) and is known to regulate motor output (Smith et al., 1998; Kita, 2007; Hernández et al., 2015; Glajch et al., 2016; Hegeman et al., 2016; Pamukcu et al., 2020). Consistent with this idea, decorrelated, phasic changes in GPe neuron activity are observed with normal movements (Anderson and Horak, 1985; Shi et al., 2004; Turner and Anderson, 2005; Jin et al., 2014; Dodson et al., 2015; Mallet et al., 2016). Alterations in the firing pattern of these neurons are associated with hypokinetic motor symptoms in both animal models of Parkinson's disease and human patients (Filion et al., 1991; Hutchison et al., 1994; Nini et al., 1995; Rothblat and Schneider, 1995; Boraud et al., 1998; Raz et al., 2000; Magill et al., 2001; Mallet et al., 2008; Chan et al., 2011; Jaeger and Kita, 2011).
Although the importance of the GPe has long been recognized, our understanding of its cell types, their organization, and their functional properties is limited. While prior studies argue that GPe neurons are involved in movement control (Dodson et al., 2015; Zimnik et al., 2015; Glajch et al., 2016; Mastro et al., 2017; Aristieta et al., 2020; Gu et al., 2020; Pamukcu et al., 2020), precisely how they are involved in motor function and dysfunction remains poorly defined. A major hurdle has been imposed by the complexity of the cellular makeup of the GPe. Our past work characterized two principal classes of GPe neurons, parvalbumin-expressing (PV+) neurons and Npas1-expressing (Npas1+) neurons that account for ∼50% and ∼30% of the GPe, respectively (Hernández et al., 2015; Hegeman et al., 2016; Abecassis et al., 2020). Aside from Foxp2-expressing (Foxp2+) neurons that form a unique subclass within the Npas1+ class, we lack driver lines that selectively capture other GPe neuron subclasses. The advent of transcriptomic data provided key insights for genetic dissection of GPe neuron subtypes. In particular, preexisting single-cell transcriptomic studies (Saunders et al., 2018) and in situ hybridization data (Lein et al., 2007) suggest the existence of molecular markers that can be used but have not been previously explored for the interrogation of GPe neuron subtypes. In this study, we examined the utility of Kcng4-Cre, Npr3-Cre, and Npy2r-Cre mouse lines. Using complementary approaches, such as anatomical, electrophysiological, and behavioral approaches, we concluded that Npr3+ neurons and Kcng4+ neurons are unique neuron subclasses within the GPe. Critically, capitalizing on newly available transgenic tools, we showed that GPe neuron subtypes were distinct in their roles in regulating motor patterns.
Materials and Methods
Mice
All procedures were done in accordance with protocols approved by Northwestern University and the University of Texas Health Science Center at Houston Institutional Animal Care and Use Committees and were in compliance with the National Institutes of Health's Guide for the care and use of laboratory animals. Experiments were conducted with the following mouse lines: Adora2a-Cre BAC (A2a-Cre, MMRRC 031168), C57BL/6J (C57, Jax 000664), Drd1a-Cre BAC (D1-Cre, MMRRC 029178), Foxp2-ires-Cre (Foxp2-Cre, Jax 030541), FSF(Frt-STOP-Frt)-LSL-tdTomato (Ai65, Jax 021875), Kcng4-Cre (Jax 029414), Lox-STOP-Lox(LSL)-tdTomato (Ai14, Jax 007914), Npr3-ires2-Cre (Npr3-Cre, Jax 031333), Npy2r-ires-Cre (Npy2r-Cre, Jax 029285), Npas1-Cre-tdTomato BAC (Npas1-Cre, Jax 027718), PV-ires-Cre (PV-Cre, Jax 017320), PV-2A-Flp (PV-Flp, Jax 022730), PV-tdTomato BAC (PV-tdTomato, Jax 027395). To genetically label different neuron populations for histological and electrophysiological studies, the following crosses were made: Foxp2-Cre;PV-tdTomato, Npr3-Cre;Ai14 (Npr3-L-tdTomato), Npy2r-Cre;Ai14 (Npy2r-L-tdTomato), PV-Flp;Npy2r-Cre;Ai65 (PV-Npy2r-FL-tdTomato), PV-Cre;Ai14 (PV-L-tdTomato), PV-tdTomato;Lhx6-eGFP, Npas1-Cre;Lhx6-eGFP, and Npas1-Cre;PV-tdTomato. Only heterozygous and hemizygous mice were used throughout the study to minimize the potential alteration of the phenotypes in mice carrying the transgene alleles (Chan et al., 2012). All mouse lines were maintained by backcrossing with C57BL/6J stock. Mice were group-housed in a 12 h light-dark cycle. Food and water were provided ad libitum. The genotypes of all transgenic mice were determined by tail biopsy followed by PCR to identify the presence of the relevant transgenes. Both male and female mice were used in this study.
Immunolabeling and biocytin visualization
Mice ranging in age from postnatal day 55–80 were anesthetized deeply with a ketamine-xylazine mixture and perfused transcardially first with phosphate-buffered saline (PBS) followed by a fixative containing 4% (w/v) paraformaldehyde, pH 7.4. Tissue was then postfixed in the same fixative for 2 h at 4°C. Tissue blocks containing the GPe were sectioned using a vibrating microtome (Leica Instruments) at a thickness of 60 μm. Floating sections were blocked with 10% (v/v) normal goat or donkey serum (Thermo Fisher Scientific) and 0.2% (v/v) Triton X-100 in PBS for 30–60 min, and were subsequently incubated with primary antibodies in the same solution for 16–24 h at 4°C. Details of the primary antibodies used in this study are listed in Table 1. After washes in PBS, the sections were incubated with Alexa-conjugated secondary antibodies (Thermo Fisher Scientific, 1:500 dilution) at room temperature for 2 h. The sections were then washed, mounted with ProLong Antifade mounting medium (Thermo Fisher Scientific), and coverslipped. In a subset of the experiments, DAPI was used to delineate cytoarchitecture of different brain structures. Fluorescent images of injection sites were captured on an epifluorescence microscope (Keyence) using a 2 × or 10 × 0.45 numerical aperture (NA) objective. Immunoreactivity in neurons was examined on a laser-scanning confocal microscope (Olympus). For cell quantification, images of the entire GPe were acquired on a laser-scanning confocal microscope with a 60 × 1.35 NA oil-immersion objective. Images encompassing the GPe were taken and stitched using FLUOVIEW Viewer (Olympus) or Photoshop (Adobe Systems). Cell counting was performed manually using the cell-counter plugin within Fiji (Schindelin et al., 2012). Cell counts were obtained from optical sections that were captured at 1 μm increments. Neurons were defined by cells that were immuno-positive for HuCD or NeuN (Hernández et al., 2015; Abecassis et al., 2020). GPe sections from three different equally-spaced (400 μm) mediolateral levels (∼2.5, 2.1, and 1.7 mm from bregma) were sampled and assigned as lateral, intermediate, and medial, respectively (Hernández et al., 2015; Abecassis et al., 2020). They correspond approximately to sagittal plates 7, 9, and 11 on the Allen reference atlas (http://mouse.brain-map.org/static/atlas). In this study, the GPe is considered to be the structure that spans between the dorsal striatum and the internal capsule, which define the rostral and caudal borders of the GPe on a sagittal plane, respectively. The cytoarchitecture of the ventral border is more ambiguous. For consistency, six nonoverlapping z stacks (212.13 × 212.13 μm) traversing the long axis of the GPe were used to capture its dorsoventral extent.
Table 1.
Primary antibodies used in this study
| Antigen | Host species | Clonality | Source | Catalog no. | Lot no. | Dilution | Working concentration |
|---|---|---|---|---|---|---|---|
| Foxp2 | Rabbit | Polyclonal | Sigma Millipore | HPA000382 | B115858 | 1:500 | — |
| GFP | Chicken | Polyclonal | Abcam | ab13970 | GR3190550-9 | 1:1000 | — |
| HuC/D | Mouse | Monoclonal | Invitrogen | A21271 | 1900217 | 1:1000 | 0.2 μg/ml |
| NeuN | Rabbit | Polyclonal | Biosensis | R-3770-100 | R-3770-300-201605-SH | 1:1000 | 1.0 μg/ml |
| Nkx2.1 | Rabbit | Polyclonal | Sigma Millipore | 07-601 | 2887266 | 1:500 | — |
| Npas1a | Guinea pig | Polyclonal | — | — | — | 1:5000 | — |
| PV | Guinea pig | Polyclonal | Synaptic Systems | 195004 | 195004/1-19 | 1:1000–1:2000 | — |
| RFP | Mouse | Monoclonal | Rockland | 200-301-379 | 34537 | 1:1000 | 0.5 μg/ml |
| tdTomato | Rat | Monoclonal | Kerafast | EST203 | 091918 | 1:1000 | 0.5 μg/ml |
aSee Hernández et al. (2015).
To visualize biocytin GPe neurons, 0.2% (w/v) biocytin was included in the internal solution and recorded in the whole-cell mode for at least 30 min (see below). Recovered slices were flat-mounted on nitrocellulose filters (Millipore) and fixed overnight in 4% (w/v) paraformaldehyde, pH 7.4, at 4°C. After several washes in PBS, slices were reacted 3 times in freshly prepared 0.5% (w/v) NaBH4 in H2O. Slices were then incubated in fresh Scale (Hama et al., 2011) solution (4 m urea, 10% (v/v) glycerol and 0.1% Triton X-100) for 10 d at 4°C. Subsequently, slices were washed in PBST (1% Triton X-100 in PBS) and blocked in 10% (v/v) normal goat serum and PBST for 45 min. After several washes, slices were reacted with 2 μg/ml streptavidin-AlexaFluor-647 (Thermo Fisher Scientific) in blocking solution overnight at 4°C in the dark. Slices were washed in PBS followed by Scale solution. Sections were mounted, air-dried, and then coverslipped in ProLong Gold mounting medium (Thermo Fisher Scientific). Serial optical sections (Z stacks) were acquired on a laser-scanning confocal microscope (Olympus) with a 60 × 1.35 NA oil-immersion objective (Olympus) at 1 μm intervals and stitched together using Fiji.
Stereotaxic injections and fiber implantations
For in vivo optogenetic experiments, mice aged postnatal day 28–35 were anesthetized in an isoflurane induction chamber at 3%–4% isoflurane and immobilized on a stereotaxic frame (David Kopf Instruments). Anesthesia was maintained using 1%–2% isoflurane. The scalp was opened using a scalpel and a small craniotomy (1 mm diameter) was made with a dental drill (Osada). Adeno-associated viruses were infused with calibrated 5 μl glass pipettes (VWR) pulled to have a tip diameter of 3 μm. The injection needle was left in situ for 5–10 min following the end of the injection to maximize tissue retention of adeno-associated viruses and decrease capillary spread on pipette withdrawal. Experiments were performed 4–6 weeks after stereotaxic surgeries. The accuracy of injections was visually inspected under epifluorescence microscopy in ex vivo slices or histologically verified post hoc.
To allow for optogenetic manipulations, fiber optic cannulae were implanted bilaterally at the region of interest (Pamukcu et al., 2020) 3 weeks after stereotaxic injections. Each fiber cannula was assembled with an optical fiber (0.66 NA, 250 μm core) (Prizmatix) secured into a zirconia ferrule (1.25 mm OD) (Prizmatix) with nonfluorescent epoxy (Thorlabs). To determine the intensity of light exiting fibers, the output of fibers was measured with a power meter (Thorlabs). Cannulae were fixed to the skull using dental cement (Parkell).
Behavioral testing
In vivo optogenetic interrogation was performed 1–2 weeks after fiber implantations. Behavioral tests were performed between 2:00 P.M. and 7:00 P.M. In vivo optogenetic experiments were performed in an opaque white plastic open-field box (28 cm × 28 cm), which was cleaned with 70% ethanol. On the day before optogenetic experiments, each mouse was allowed to acclimate to the open-field box for 20 min, with a fiber optic cable attached to the implant via a rotary joint (Prizmatix). Pre-trial, basal ambulatory activity was collected for 5 min at the beginning of the behavioral session. For ChR2 activation, mice were subjected to either a single, sustained light pulse or a patterned pulse train (5 ms pulses at 20 Hz) delivered for 10 s. For GtACR2 activation, only the sustained protocol was used. A blue (peak, ∼455 nm) LED (Prizmatix) was used to activate both opsins. The light power used for opsin activation was 12–18 mW measured at the fiber tip. Stimuli were delivered with a 1 min intertrial interval. An overhead camera (Logitech) was used to acquire mouse behavior in the open field arena at 30 fps, 640 pixels × 240 pixels per frame. Videos were subsequently cropped and downsampled to 10 fps at full resolution for markerless tracking.
Behavioral tracking and classification
DeepLabCut (https://github.com/DeepLabCut/) (Mathis et al., 2018; Nath et al., 2019) was used for tracking body parts of mice in an open field arena. Eight body parts, including the nose, ears, body center, side laterals (hip joints), tail base, and tail end, were labeled in top-down view videos (see Fig. 5a). To create the training dataset, 1674 distinct frames from 50 video recordings of open-field behavior were manually annotated. We used MobileNetV2-1-based network (Sandler et al., 2018; Mathis et al., 2019) with default parameters. The network was trained and refined for five rounds using default multistep learning rates. Each round consists of 240,000–1,000,000 iterations, and the default multistep learning rates were used. This trained network has a test error of 4.82 pixels and a training error of 1.13 pixels. Predictions of X-Y coordinates were processed using a median filter with a rolling window of five frames before further analysis. To cross-validate our network performance, speeds computed from DeepLabCut and Ethovision data were compared; near-identical results were obtained (see Results). This network was then used to analyze all videos in this study.
Figure 5.
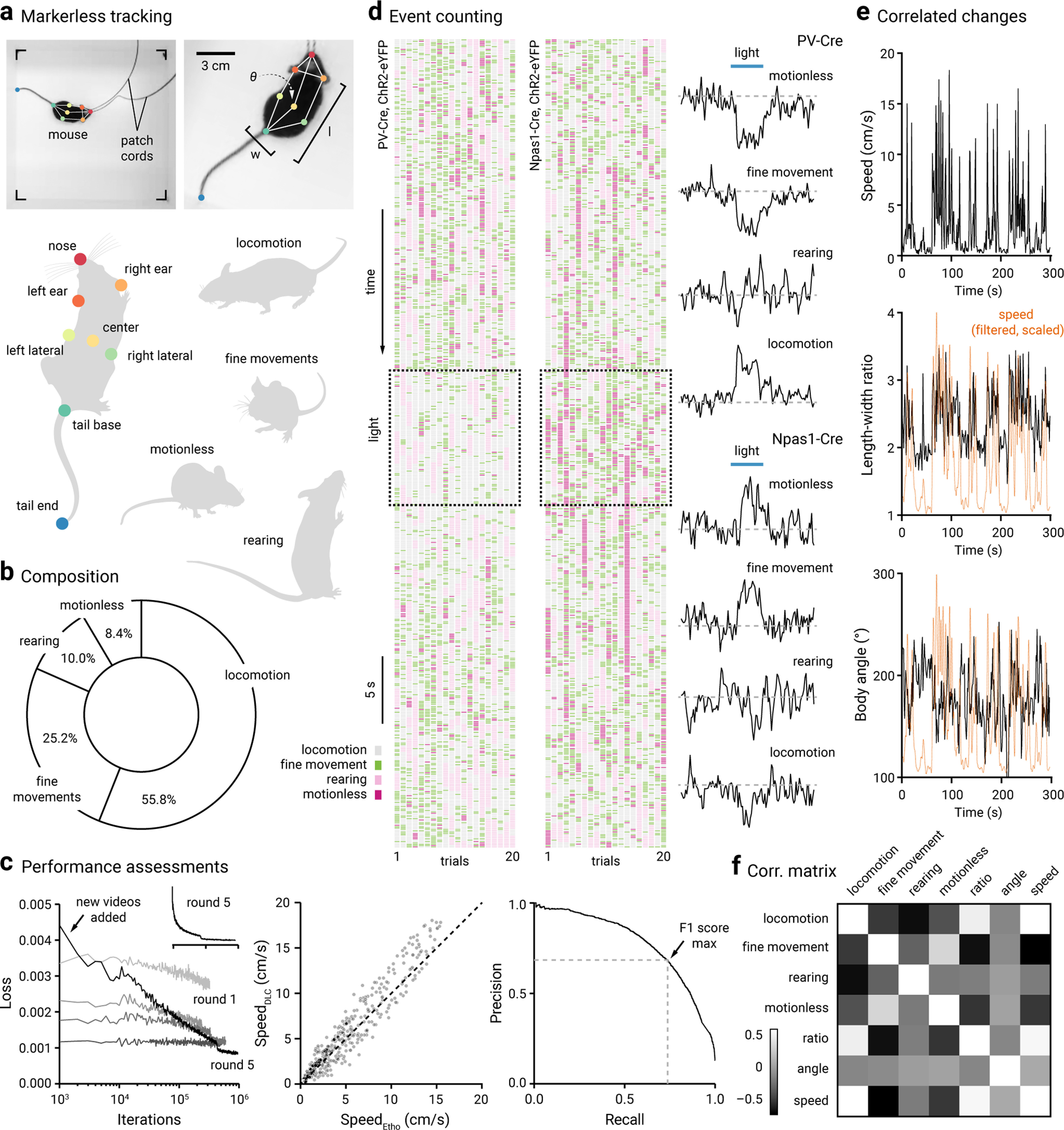
Markerless tracking and behavioral dynamics. a, Top left, A still frame showing a mouse exploring in an open-field arena. Patch cords were attached to the fiber implants for optogenetic interrogation. Color circles represent pose estimation markers. Top right, Magnified view of the same mouse shown on the left. Eight body parts were labeled and used for tracking body kinematics and motor patterns. Angle (θ), width (w), and length (l) were measured. Bottom, A schematized labeling configuration is shown. Right, Four discrete motor behavior motifs were extracted, namely, locomotion, fine movement, motionless, and rearing. b, The behavioral composition is shown as percent time spent. PV-Cre (n = 12) and Npas1-Cre (n = 16) mice virally transduced with ChR2-eYFP were included in this analysis. Mice were tethered to the patch cords but no light was delivered. c, Left, Learning curves of the DeepLabCut network over five rounds of training. Additional annotated frames were incorporated into the dataset in each round. An initial steep and monotonic decrease in loss was followed by a steady plateau. A large amount of new data were added to Round 5, resulting in a high loss initially. The same data from Round 5 were replotted on a linear scale as an Inset (top right). Middle, An X-Y scatter plot comparing the speeds measured using DeepLabCut versus Ethovision. Each marker is a frame. Diagonal line indicates unity (i.e., x = y). Right, Precision-recall curve of the SimBA rearing classifier. Arrow indicates the maxima for the F1 score (0.71). d, Left and Middle, Ethograms showing the motor dynamics of a PV-Cre mouse (left) and Npas1-Cre mouse (right) across 20 1 min trials (columns). The beginning of each trial starts at the top of the plots. Square boxes represent the time period in which light was delivered in the GPe. Rasters represent the occurrence of defined behavioral events: locomotion (gray), fine movement (green), rearing (light pink), and motionless (magenta). Each column is a single trial; 20 trials are shown for both PV-Cre and Npas1-Cre. Right, Population average changes in the event frequency of motionless, fine movement, rearing, and locomotion upon optogenetic stimulation (blue horizontal lines) of PV-Cre (n = 12, top) and Npas1-Cre (n = 16, bottom) mice transduced with ChR2-eYFP. e, Top to bottom, Correlated changes in speed, length-width ratio, and body angle of an example mouse across time. Speed data (orange dotted lines) were filtered and scaled to facilitate comparisons. f, A correlation matrix constructed using time series data from both pre period and light period illustrating moment-to-moment covariance of the different movement metrics. The event frequencies of the four behaviors, body angle, length-width ratio, and speed were used in this comparison. PV-Cre and Npas1-Cre mice (virally transduced with ChR2-eYFP) were included in this analysis (n = 28). Light colors represent positive correlations. Dark colors represent negative correlations.
To categorize motor behavior, DeepLabCut tracking data were first calibrated; the pixel-to-centimeter conversion for each video was determined by comparing the width of the arena in pixels to the actual width of the arena (28 cm). Based on the calibrated X-Y coordinates of labeled body parts, a set of movement metrics was generated for each frame. Mouse speed was measured as the body center speed. Body angle was defined as tail base-nose angle relative to the body center. An angle <180° indicates the body axis is skewed to the right, and an angle >180° indicates the body axis is skewed to the left. Mouse width was measured as the Euclidean distance between the side laterals, and mouse length was measured as the Euclidean distance between the nose and the tail base. Locomotion was defined as frames when the body center had a speed >0.5 cm/s; motionless was defined as frames when the ears, body center, laterals, and tail base all had a speed ≤0.5 cm/s. To classify rearing, we constructed a random forest classifier in SimBA; 19,058 rearing frames from 35 video recordings of open-field behavior were extracted and manually annotated as rearing by three independent annotators. Supported and unsupported rearing behaviors were not differentiated. The start frame was defined as the frame in which the mouse lifted its forelimbs off the floor and extended its head upwards; the end frame was defined as the frame before the forelimbs made contact with the floor. The model was built with the following settings: n_estimators = 2500, RF_criterion = entropy, RF_max_features = sqrt, RF_min_sample leaf = 2, and no oversampling or undersampling; 20% of the video frames were used for testing and the other 80% were used for training. The resulting classifier has a F1 score = 0.71, precision = 0.68, and recall = 0.74. The performance of this classifier was on par with those reported recently (Nilsson et al., 2020). The discrimination threshold was set at Pr = 0.31, and each instance of rearing had a minimum duration of 300 ms. As neighboring frames are highly correlated, the random test-train splitting approach may result in an overestimation of performance. To this end, we built a classifier with the same dataset using a time series split: the first 80% of the frames were used for training and the last 20% of the frames were used for testing. This classifier yielded F1 score = 0.67, precision = 0.66, and recall = 0.67. Last, fine movement was defined as frames that did not fall into any of the categories mentioned above (i.e., locomotion, motionless, or rearing). Finally, example videos and the trained model are available on Github (https://github.com/saviochan/SimBA-OpenFieldArena, https://github.com/saviochan/Deeplabcut-OpenFieldArena) and Zenodo (https://zenodo.org/record/3964701#.XyB8yJ5KhPZ, https://zenodo.org/record/4556331#.YDceFegzZnI). The data generated by the analysis pipeline were processed using custom Python scripts. Codes are available online (https://github.com/saviochan/Python-Scripts/tree/master/OpenFieldArena_Behavior). Twenty-five different movement metrics were tracked. Event frequency, duration, and percent time spent were logged. Event frequency is the average of the total event counts in 10 s across all trials. “Light-period” corresponds to 10 s of light delivery. “Pre-period” and “post-period” correspond to the 10 s epoch before and after light delivery, respectively. Fold changes were calculated by dividing the movement metric during light period by that in pre period.
To assess the relationship between the measured movement metrics, a correlation matrix was constructed from binned, time-series data. Rearing-motionless switch frequency and motionless-rearing switch frequency were excluded because of the low occurrence of events. Hierarchical clustering of movement metrics was performed in ClustVis (https://biit.cs.ut.ee/clustvis/) (Metsalu and Vilo, 2015). Twenty-five movement metrics were included in the analysis. Mice with targeted optogenetic stimulation of Foxp2+ neurons, Kcng4+ neurons, Npas1+ neurons, and PV+ neurons were included. Both rows and columns were clustered using correlation distance and average linkage. Movement metrics were centered and scaled. A K-nearest neighbors classification algorithm was implemented in JASP to construct a decision boundary matrix. All variables were scaled to have a mean of 0 and an SD of 1. The data were split as follows: 60% training, 20% testing, and 20% validation.
Visualized ex vivo electrophysiology
Mice in the age range postnatal day 55–100 were anesthetized with a ketamine-xylazine mixture and perfused transcardially with ice-cold aCSF containing the following (in mm): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 2.0 CaCl2, 1.0 MgCl2, 25 NaHCO3, and 12.5 glucose, bubbled continuously with carbogen (95% O2 and 5% CO2). The brains were rapidly removed, glued to the stage of a vibrating microtome (Leica Instrument), and immersed in ice-cold aCSF. Parasagittal slices containing the GPe were cut at a thickness of 240 μm and transferred to a holding chamber where they were submerged in aCSF at 37°C for 30 min and returned to room temperature for recording. Slices were then transferred to a small-volume (∼0.5 ml) Delrin recording chamber that was mounted on a fixed-stage, upright microscope (Olympus). Neurons were visualized using differential interference contrast optics (Olympus), illuminated at 735 nm (Thorlabs), and imaged with a 60× water-immersion objective (Olympus) and a CCD camera (QImaging). Genetically-defined neurons were identified by somatic eGFP or tdTomato fluorescence examined under epifluorescence microscopy with a daylight (6500 K) LED (Thorlabs) and appropriate filters (Semrock).
Recordings were made at room temperature (20°C–22°C) with patch electrodes fabricated from capillary glass (Sutter Instrument) pulled on a Flaming-Brown puller (Sutter Instrument) and fire-polished with a microforge (Narishige) immediately before use. Pipette resistance was typically ∼3–5 MΩ. For cell-attached and current-clamp recordings, the internal solution consisted of the following (in mm): 135 KMeSO4, 10 Na2phosphocreatine, 5 KCl, 5 EGTA, 5 HEPES, 2 Mg2ATP, 0.5 CaCl2, and 0.5 Na3GTP, with pH adjusted to 7.25–7.30 with KOH. For voltage-clamp recordings of inhibitory postsynaptic currents (IPSCs), a high-chloride internal solution consisted of the following (in mm): 120 CsCl, 10 Na2phosphocreatine, 5 HEPES, 5 tetraethylammonium-Cl, 2 Mg2ATP, 1 QX314-Cl, 0.5 Na3GTP, 0.5 CaCl2, 0.25 EGTA, and 0.2% (wt/vol) biocytin, pH adjusted to 7.25–7.30 with CsOH and a low-chloride internal solution consisted of the following (in mm): 125 CsMeSO3, 10 Na2-phosphocreatine, 5 tetraethylammonium chloride, 5 QX-314 Cl, 5 HEPES-K, 5 EGTA-K, 2 Mg2ATP, 0.5 CaCl2, 0.5 Na3GTP, 0.2% (w/v) biocytin, with pH adjusted to 7.25–7.30 with CsOH. Stimulus generation and data acquisition were performed using an amplifier (Molecular Devices), a digitizer (Molecular Devices), and pClamp (Molecular Devices). For current-clamp recordings, the amplifier bridge circuit was adjusted to compensate for electrode resistance and was subsequently monitored. The signals were filtered at 1 kHz and digitized at 10 kHz. KMeSO4 and Na2-GTP were from ICN Biomedicals and Roche, respectively. All other reagents were obtained from Sigma Millipore.
For optogenetic experiments, blue excitation wavelength (peak, ∼450 nm) from two daylight (6500 K) LEDs (Thorlabs) was delivered to the tissue slice from both a 60× water immersion objective and a 0.9 NA air condenser with the aid of 520 nm dichroic beamsplitters (Semrock). Light delivery was made at the site of electrophysiological recordings with a field of illumination of 500–700 μm in diameter. Paired-pulse optogenetic activation of terminals was at 20 Hz with a light duration of 2 ms. To extract the properties of spontaneous IPSCs in identified GPe neurons, an analysis similar to that described by Higgs and Wilson (2016) was used. Briefly, an autocorrelation of spontaneous IPSC times was computed; and for each periodic component detected, the periodic peaks were fitted with a model curve to estimate the presynaptic firing frequency, the coefficient of variation (CV) of presynaptic interspike intervals (ISIs), and the synaptic success probability. The unitary IPSC amplitudes and waveforms were estimated by subdividing spontaneous IPSCs based on their timing with respect to the surrounding periodic input.
The electrophysiological data used in this study were expanded from a previously published dataset (Abecassis et al., 2020). Hierarchical clustering of electrophysiological attributes was performed in ClustVis (https://biit.cs.ut.ee/clustvis/) (Metsalu and Vilo, 2015). All cell-attached and whole-cell measurements were included in this analysis. Both rows and columns were clustered using correlation distance and average linkage. Electrophysiological measurements were centered and scaled. Principal component analysis (PCA) of electrophysiological attributes was performed using the R programming language. t-SNE (k = 2) was performed in JASP using the Hartigan-Wong algorithm with the exact same dataset that was used for hierarchical clustering. All variables were scaled. Partial correlation networks based on Pearson coefficients of electrophysiological parameters were analyzed in JASP and Cytoscape (Shannon et al., 2003) with the CentiScaPe plug-in (Scardoni et al., 2009). The complete dataset for electrophysiological measurements of the intrinsic properties and an interactive web tool can be found online (https://icbi-georgetown.shinyapps.io/Neuro_interactive_analysis/).
Drugs
R-CPP and NBQX disodium salt were obtained from Tocris Bioscience. CGP55845 and QX314-Cl were obtained from Abcam. Na3GTP and tetrodotoxin were from Roche and Alomone Laboratories, respectively. Other reagents not listed above were from Sigma Millipore. Drugs were dissolved as stock solutions in either water or DMSO and aliquoted and frozen at –30°C before use. Each drug was diluted to the appropriate concentrations by adding to the perfusate immediately before the experiment. The final concentration of DMSO in the perfusate was < 0.1%.
Experimental design and statistical analyses
General graphing and statistical analyses were performed with MATLAB (The MathWorks), Prism (GraphPad), JASP (https://jasp-stats.org), and the R programming language (https://www.r-project.org). Custom analysis codes are available on GitHub (https://github.com/saviochan). Sample size (n value) is defined by the number of observations (i.e., neurons, sections, or mice). When percentages are presented, n values represent only positive observations. No statistical method was used to predetermine sample size. Data in the main text are presented as median values ± median absolute deviations (MADs) (Leys et al., 2013) as measures of central tendency and statistical dispersion, respectively. Box plots are used for graphic representation of population data unless stated otherwise (Krzywinski and Altman, 2014; Streit and Gehlenborg, 2014; Nuzzo, 2016). The central line represents the median, the box edges represent the interquartile ranges, and the whiskers represent 10–90th percentiles. Normal distributions of data were not assumed. Individual data points were visualized for small sizes or to emphasize variability in the datasets. Nonparametric statistics were used throughout. Comparisons of unrelated samples were performed using a Mann–Whitney U test, and comparisons of paired samples were performed using the Wilcoxon signed rank test. The Kruskal–Wallis test was used to compare multiple groups, followed by the post hoc Dunn test to assess pairwise differences while adjusting for the number of pairwise comparisons. The Spearman exact test was used for evaluating correlation between variables with a threshold (α) of 0.05 for significance. Unless < 0.0001 or > 0.99, exact p values (two-tailed) are reported in the text. To avoid arbitrary cutoffs and visual clutter, levels of significance are not included in the figures. Statistical significance is assessed at a significance level of 0.05, after applying the Bonferroni correction to adjust for the number of comparisons.
Results
Recent single-cell transcriptomic analyses have revealed four distinct neuron clusters within the GPe (Saunders et al., 2018). In addition to the well-established Npas1+-Foxp2+ neurons (blue) that form a subclass of Npas1+ neurons, Npas1+-Nkx2.1+ neurons (cyan) are the other subclass of Npas1+ neurons (Fig. 1a). Furthermore, PV+ neurons can be subdivided into two clusters (red and teal) with differential expression of Npr3, Kcng4, and Npy2r. Raw data showing the expression patterns of Npr3, Kcng4, and Npy2r can be found online (http://dropviz.org/?_state_id_=6c13807c8420bc74). The expression of these markers in the adult GPe is in agreement with results from in situ hybridization (Fig. 1b). While sparse signals were found with Npr3 and Kcng4, the signals associated with Npy2r were widespread through the GPe. Here, we sought to examine whether Npr3-Cre (Daigle et al., 2018), Kcng4-Cre (Duan et al., 2014), and Npy2r-Cre (Chang et al., 2015) mice could provide genetic access to unique GPe subpopulations.
Figure 1.
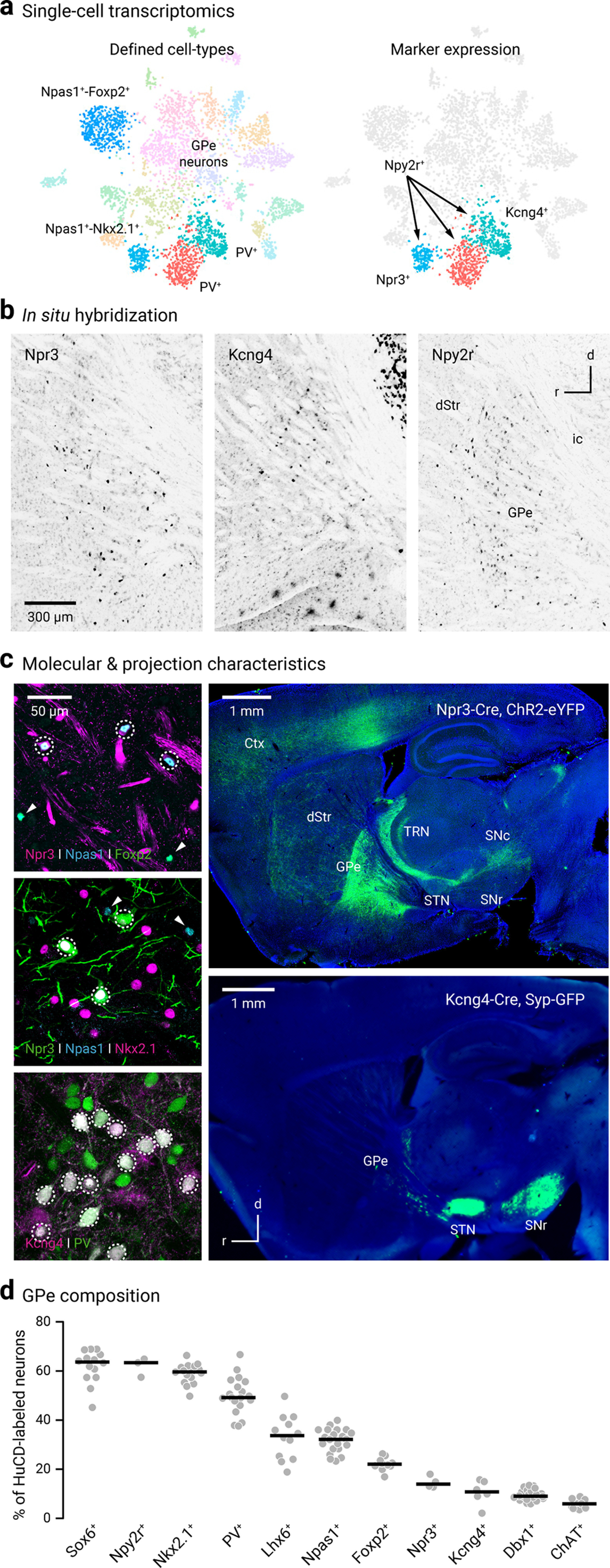
Novel markers and driver lines for interrogating GPe neurons. a, Left, Single-cell transcriptomic data identified four distinct clusters of GPe neurons; these include Npas1+-Foxp2+ (blue, 10 o'clock), Npas1+-Nkx2.1+ (cyan, 7 o'clock), and two subpopulations of PV+ neurons (red and teal, 6 o'clock). Right, Npy2r is expressed in all non-Npas1+-Foxp2+ neurons; Npr3 and Kcng4 are expressed in a select subset of Npas1+ neurons and PV+ neurons, respectively. Adapted from Saunders et al. (2018). Original expression data can be viewed at http://dropviz.org/?_state_id_=6c13807c8420bc74. b, Sagittal brain sections showing the in situ hybridization signals for Npr3 (left), Kcng4 (middle), and Npy2r (right) in the external globus pallidus (GPe) and neighboring areas. Data were adapted from Allen Brain Atlas. Brightness and contrast were adjusted. Raw data can be viewed and downloaded from http://mouse.brain-map.org. dStr, dorsal striatum; ic, internal capsule. c, Top left, Coexpression (dotted circle) of Npas1 (blue) but not Foxp2 (green) in Npr3+ neurons (magenta). Npas1+-Foxp2+ (and non-Npr3+) neurons were visible in the same field (arrowheads). Middle left, Coexpression (dotted circle) of Npas1 (blue) and Nkx2.1 (magenta) in Npr3+ neurons (green). Npas1+ neurons that were neither Nkx2.1+ nor Npr3+ were visible in the same field (arrowheads). Bottom left, Coexpression (dotted circle) of PV (green) in Kcng4+ neurons (magenta). Top right, Npr3+ neurons share axonal projection patterns similar to that of Npas1+-Nkx2.1+; strong axonal projections to the cortex (Ctx), thalamic reticular nucleus (TRN), and substantia nigra pars compacta (SNc) were evident. Projections were assessed using viral delivery of ChR2-eYFP. Bottom right, Virally delivered synaptophysin-GFP in Kcng4-Cre mice revealed near-exclusive axonal projection from the GPe to the subthalamic nucleus (STN) and substantia nigra pars reticulata (SNr). d, Using HuCD as a neuronal marker, population data for the relative abundance of GPe neuron markers were determined. Each circle represents a section. Medians and sample sizes are as follows: Sox6+ (63.7%, 14), Npy2r+ (63.4%, 3), Nkx2.1+ (59.6%, 16), PV+ (49.2%, 19), Lhx6+ (33.7%, 12), Npas1+ (32.2%, 21), Foxp2+ (22.1%, 10), Npr3+ (14.0%, 4), Kcng4+ (10.8%, 6), Dbx1+ (9.0%, 22), ChAT+ (5.9%, 9). Data are ranked based on the median values as denoted by the thick lines. Parts of the data have been previously published.
Novel driver lines capture distinct neuron populations
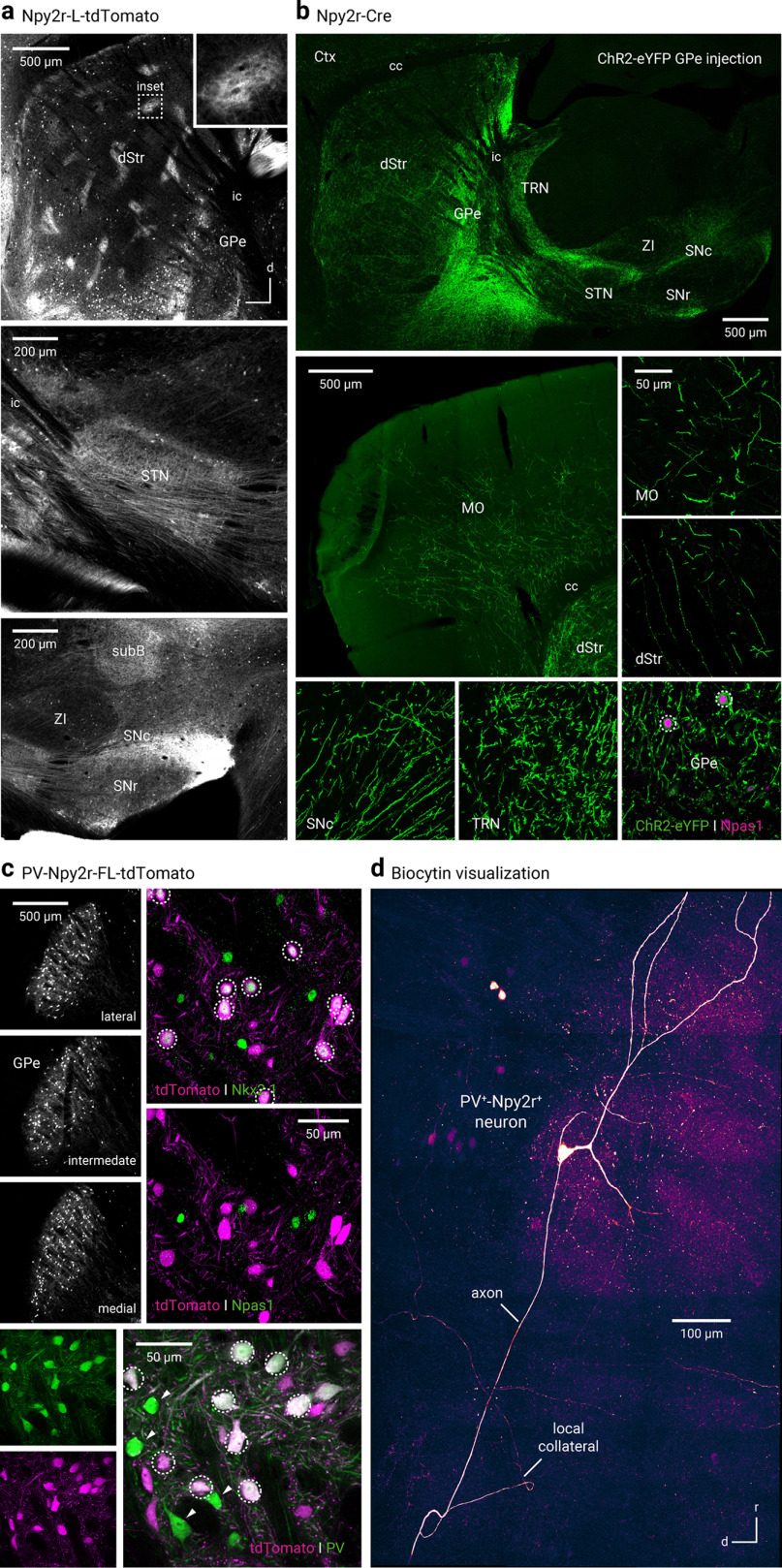
To study Npr3+ neurons, tdTomato-expressing (tdTomato+) neurons in Npr3-L-tdTomato mice were used. Npr3+ neurons were 14% of GPe neurons (14 ± 1%, n = 3 sections, 270 neurons); ∼90% of Npr3+ neurons were Npas1+ (89 ± 4%, n = 4 sections, 474 neurons), 74% were Nkx2.1+ (74 ± 0%, n = 3 sections, 169 neurons), and 2% were Foxp2+ (2 of 170 neurons) (Fig. 1d). To visualize the axonal projection pattern of Npr3+ neurons, ChR2-eYFP was transduced. This resulted in clear axonal arborization within the GPe, in addition to strong eYFP signals in the thalamic reticular nucleus, cortex, and substantia nigra pars compacta (SNc; Fig. 1c). Our data are in line with the recent report that Lhx6+ GPe neurons strongly target compacta neurons (Evans et al., 2020). In summary, Npr3+ neurons had molecular and projectional properties that were completely consistent with those of Npas1+-Nkx2.1+ neurons (Abecassis et al., 2020). These findings further the idea that Npas1+-Nkx2.1+ neurons, along with the previously identified Npas1+-Foxp2+ neurons, are unique GPe neuron types and corroborate the findings in our recent study (Abecassis et al., 2020).
In addition to the robust expression in the thalamic reticular nucleus, in situ hybridization suggested that Kcng4 is expressed in a subset of neurons in the GPe. To further examine the molecular properties of Kcng4+ neurons, we performed immunohistological studies using the Kcng4-L-tdTomato genetic cross. As shown in Figure 1c, the majority of Kcng4+ neurons (tdTomato+) express PV (90 ± 10%, n = 3 sections, 121 neurons), and they represent ∼30% (28 ± 8%, n = 3 sections, 121 neurons) of the PV+ population and 10% (10 ± 6%, n = 3 sections, 131 neurons) of the entirety of the GPe. These data are consistent with the single-cell transcriptomic data (Saunders et al., 2018) and show that Kcng4+ neurons are a subtype of PV+ neurons in the GPe. Using a Cre-inducible synaptophysin-GFP virus, we uncovered that Kcng4+ neurons send axonal projections to STN, internal globus pallidus, and substantia nigra pars reticulata (SNr). No additional projection targets were observed (Fig. 1c).
To label Npy2r+ neurons, a similar genetic strategy was used (see Materials and Methods). tdTomato+ neurons from Npy2r-L-tdTomato mice were studied. As expected from the single-cell transcriptomic and in situ hybridization data, a large fraction (∼60%) of GPe neurons were tdTomato+. In addition to a population of GPe neurons, striosomes throughout the striatum were labeled in the Npy2r-L-tdTomato cross and appeared to be largely devoid of cell body labeling. The tdTomato+ neurons in the striatum were likely interneurons, based on their density (Fig. 2a).
Figure 2.
Npy2r-Cre mice capture a mixed population of GPe neurons. a, Npy2r-L-tdTomato mice labeled neurons in the GPe and striosomes in the dStr (top). Inset, Magnified view of a striosome. Prominent axons were apparent in the STN (middle) and SNc and SNr (bottom). b, Axonal projection patterns from Npy2r+ neurons are similar to that of Npas1+-Nkx2.1+ neurons and Npr3+ neurons. Only a low density of axons was observed in the STN and SNr. Bottom right, ChR2-eYFP+ neurons were largely Npas1+. c, Top left, tdTomato-expressing (PV+-Npy2r+) neurons were observed throughout the GPe in PV-Npy2r-FL-tdTomato mice. PV+-Npy2r+ neurons were immunoreactive for Nkx2.1 (top right) but not for Npas1 (middle right). Bottom, Most of PV+-Npy2r+ (tdTomato+) neurons were PV+. PV+-Npy2r– (green) neurons were visible in the same field (arrowheads). White circles represent colocalization. d, Composite confocal micrograph showing a typical biocytin-filled PV+-Npy2r+ neuron. In addition to the main axon, local collaterals were observed. cc, Corpus callosum; MO, motor cortex; SubB, subbrachial nucleus; ZI, zona incerta.
Although single-cell transcriptomic data suggest a broad expression profile of Npy2r among GPe neurons, whether the Npy2r-Cre mouse line would provide genetic access to selective neuron subtypes has not been tested. To this end, we transduced ChR2-eYFP to Cre+ neurons in Npy2r-Cre mice. This yielded axonal labeling patterns that were the most consistent with Npr3+ (also known as Npas1+-Nkx2.1+) neurons (compare Fig. 1c). In contrast, only sparse or weak axonal labeling was observed in canonical targets of PV+ neurons, i.e., STN and SNr (Fig. 2b). The unexpected results prompted us to further examine the unusual properties of the Npy2r-Cre line. As roughly half of the tdTomato+ neurons (53 ± 8%, n = 11 sections, 1319 neurons) from Npy2r-L-tdTomato mice were PV+ (not shown), we decided to selectively study PV+-Npy2r+ neurons using the PV-Npy2r-FL-tdTomato genetic cross (Fig. 2c). PV+-Npy2r+ neurons accounted for ∼65% (63 ± 4%, n = 11 sections, 1319 neurons) of the PV+ population and amounted to ∼35% (34 ± 1%, n = 3 sections, 392 neurons) of the entire GPe. The somatodendritic morphology of PV+-Npy2r+ neurons examined was also consistent with that of PV+ neurons (Fig. 2d). To our surprise, a genetic PV-Npy2r-FL-tdTomato cross yielded tdTomato expression not only in PV+ neurons but also in a very small fraction (0.89%, n = 1 of 112 neurons) of PV– neurons (Fig. 2c). One caveat of the FSF-LSL-tdTomato reporter strains is that they allow for very effective recombination, even for strains that have weak Cre activity. However, in strains that have high Cre activity, atypical expression patterns can be observed (Madisen et al., 2010).
GPe neurons cluster into two electrophysiological groups
To study the electrophysiological properties of Npr3+ neurons, Kcng4+ neurons, Npy2r+ neurons, and PV+-Npy2r+ neurons, we performed ex vivo patch-clamp recordings using near-identical procedures described previously (Hernández et al., 2015; Abecassis et al., 2020). To make sense of the results, we combined these data with an expanded version of a previously published dataset (Abecassis et al., 2020). Univariate scatter plots were used to emphasize the variability across samples. Corroborating the histological data, electrophysiological analysis showed that Npr3+ neurons and Kcng4+ neurons are a subset of the Npas1+ class and PV+ class, respectively. It is notable that Npr3+ neurons display properties that are indistinguishable from Npas1+-Lhx6+ neurons and Lhx6+bright neurons. Apart from a strong tendency of Foxp2+ neurons and PV+ neurons to be on opposite ends of the spectrum, data are largely graded (Fig. 3; Tables 2 and 3). Given that Kcng4 is an ion channel gene, it is surprising that the expression of Kcng4 did not confer unique electrophysiological properties.
Figure 3.
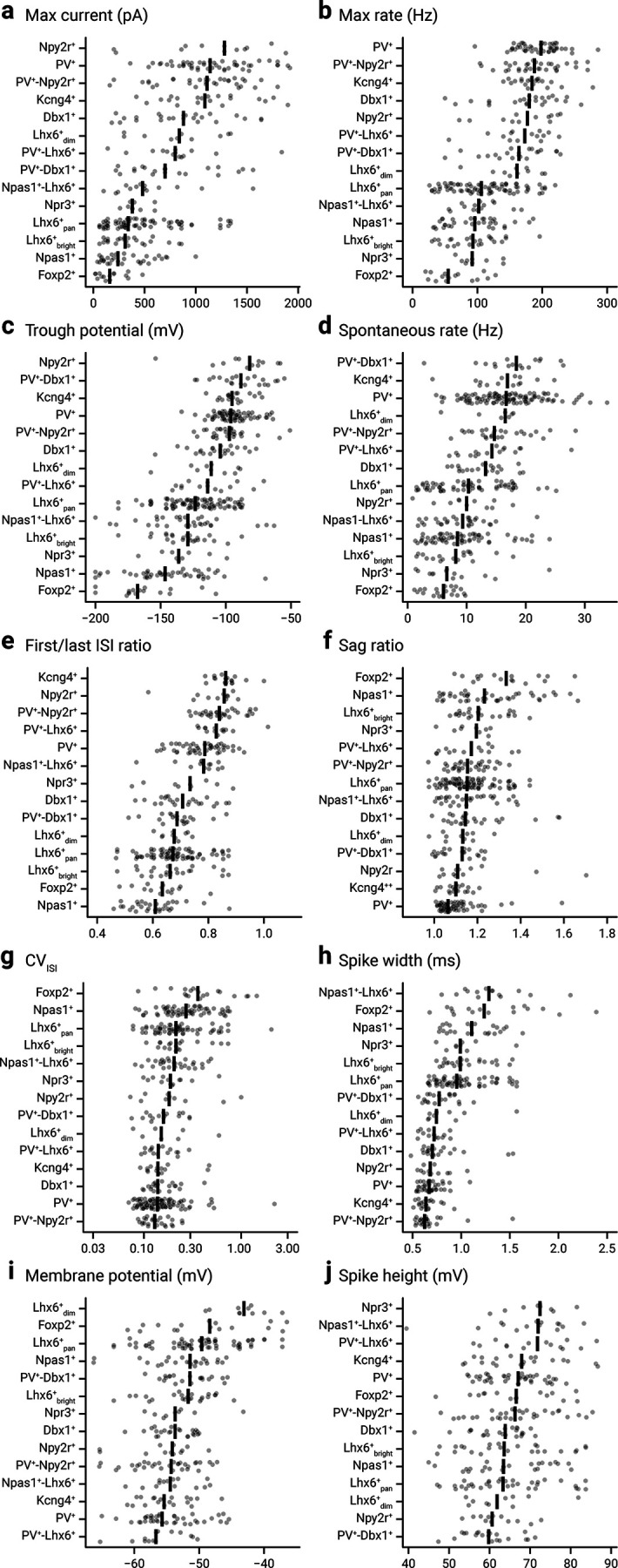
GPe neurons display graded electrophysiology attributes. Scatter plot summary of the electrophysiological properties of identified GPe neuron subtypes. Max current (a), max rate (b), trough potential (c), spontaneous rate (d), first/last ISI ratio (e), sag ratio (f), CVISI (g), spike width (h), membrane potential (i), and spike height (j) are shown. See Tables 2 and 3 for a listing of median values, sample sizes, and statistical analysis of new neuron types, compared with our recent publication (Abecassis et al., 2020), along with PV+ neurons and Npas1+ neurons for comparison. The data are ordered according to their median value as indicated by the thick vertical lines. Each cell is shown as a circle. A total of 329 neurons were included (n = Dbx1+: 24, Foxp2+: 16, Kcng4+: 20, Lhx6+bright: 24, Lhx6+dim: 10, Lhx6+pan: 68, Npas1+: 24, Npas1+-Lhx6+: 14, Npr3+: 9, Npy2r+: 12, PV+: 41, PV+-Dbx1+: 21, PV+-Lhx6+: 14, PV+-Npy2r+: 32) in this analysis. The complete dataset for electrophysiological measurements of the intrinsic properties and an interactive web tool can be found online (https://icbi-georgetown.shinyapps.io/Neuro_interactive_analysis/). CV, coefficient of variation; ISI, interspike interval.
Table 2.
Electrophysiological characteristics of GPe neurons
| Kcng4+ neurons Median ± MAD | na | Npas1+ neurons Median ± MAD | n | Npas1+-Lhx6+ neurons Median ± MAD | n | Npr3+ neurons Median ± MAD | n | Npy2r+ neurons Median ± MAD | n | PV+ neurons Median ± MAD | n | PV+-Lhx6+ neurons Median ± MAD | n | PV+-Npy2r+ neurons Median ± MAD | n | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spontaneous rate (Hz) | 16.91 ± 2.82 | 14 | 8.43 ± 3.05 | 63 | 9.31 ± 2.83 | 30 | 6.63 ± 2.51 | 13 | 10.0 ± 3.42 | 11 | 16.69 ± 3.43 | 111 | 14.26 ± 3.18 | 16 | 14.95 ± 5.59 | 22 |
| CVISI | 0.14 ± 0.03 | 14 | 0.27 ± 0.1 | 63 | 0.2 ± 0.06 | 30 | 0.19 ± 0.04 | 13 | 0.18 ± 0.08 | 11 | 0.14 ± 0.03 | 111 | 0.14 ± 0.04 | 16 | 0.13 ± 0.04 | 22 |
| Max rate (Hz) | 184.0 ± 16.0 | 20 | 96.0 ± 34.0 | 33 | 102.0 ± 22.0 | 14 | 92.0 ± 16.0 | 9 | 177.0 ± 36.0 | 12 | 198.0 ± 24.0 | 49 | 173.0 ± 24.0 | 14 | 188.0 ± 38.0 | 32 |
| Max current (pA) | 1120.0 ± 300.0 | 19 | 170.0 ± 90.0 | 24 | 460.0 ± 140.0 | 13 | 380.0 ± 40.0 | 9 | 1280.0 ± 300.0 | 12 | 1180.0 ± 260.0 | 41 | 800.0 ± 100.0 | 14 | 1100.0 ± 580.0 | 31 |
| Membrane potential (mV) | –55.41 ± 3.23 | 20 | –51.41 ± 2.96 | 24 | –54.45 ± 1.92 | 14 | –53.71 ± 1.77 | 9 | –54.16 ± 1.7 | 12 | –55.72 ± 2.56 | 41 | –56.68 ± 2.06 | 14 | –54.31 ± 5.0 | 32 |
| Trough potential (mV) | –95.27 ± 4.54 | 20 | –154.81 ± 17.26 | 24 | –129.11 ± 16.88 | 30 | –136.17 ± 7.78 | 9 | –81.67 ± 10.4 | 12 | –98.62 ± 6.35 | 41 | –113.94 ± 18.65 | 14 | –97.28 ± 16.76 | 32 |
| Sag ratio | 1.1 ± 0.03 | 20 | 1.35 ± 0.18 | 24 | 1.15 ± 0.09 | 30 | 1.2 ± 0.07 | 9 | 1.11 ± 0.06 | 12 | 1.1 ± 0.03 | 41 | 1.17 ± 0.1 | 14 | 1.15 ± 0.08 | 32 |
| First/last ISI ratio | 0.86 ± 0.02 | 20 | 0.61 ± 0.06 | 24 | 0.78 ± 0.02 | 14 | 0.73 ± 0.06 | 9 | 0.86 ± 0.03 | 12 | 0.8 ± 0.05 | 41 | 0.83 ± 0.03 | 14 | 0.84 ± 0.05 | 32 |
| Spike height (mV) | 67.93 ± 7.33 | 20 | 63.44 ± 8.52 | 24 | 71.98 ± 5.26 | 14 | 72.44 ± 3.28 | 9 | 60.63 ± 3.39 | 12 | 67.08 ± 6.68 | 41 | 71.83 ± 7.45 | 14 | 66.26 ± 12.56 | 32 |
| Spike width (ms) | 0.64 ± 0.05 | 20 | 1.11 ± 0.23 | 24 | 1.28 ± 0.17 | 14 | 0.99 ± 0.07 | 9 | 0.68 ± 0.08 | 12 | 0.67 ± 0.06 | 41 | 0.72 ± 0.1 | 14 | 0.62 ± 0.11 | 32 |
an equal to the number of neurons.
Table 3.
Statistical analysis for electrophysiological characteristicsa
| Kruskal–Wallisb | Pairwise comparisonsc |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kcng4+ vs Npas1+ | Kcng4+ vs Npas1+-Lhx6+ | Kcng4+ vs Npr3+ | Kcng4+ vs Npy2r+ | Kcng4+ vs PV+ | Kcng4+ vs PV+-Lhx6+ | Kcng4+ vs PV+-Npy2r+ | Npas1+ vs Npas1+-Lhx6+ | Npas1+ vs Npr3+ | Npas1+ vs Npy2r+ | Npas1+ vs PV+ | Npas1+ vs PV+-Lhx6+ | Npas1+ vs PV+-Npy2r+ | Npas1+-Lhx6+ vs Npr3+ | ||
| Spontaneous activity (Hz) | <0.0001 | 0.0019 | 0.0037 | 0.014 | 0.38 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | <0.0001 | 0.057 | 0.00010 | >0.99 |
| CVISI | <0.0001 | 0.014 | 0.97 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | <0.0001 | 0.011 | <0.0001 | >0.99 |
| Max rate (Hz) | <0.0001 | <0.0001 | 0.0014 | 0.0040 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | 0.0029 | <0.0001 | 0.0059 | <0.0001 | >0.99 |
| Max current (pA) | <0.0001 | <0.0001 | 0.046 | 0.019 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | <0.0001 | <0.0001 | 0.022 | <0.0001 | >0.99 |
| Membrane potential (mV) | 0.26 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | 0.38 | 0.79 | >0.99 |
| Trough potential (mV) | <0.0001 | <0.0001 | <0.0001 | 0.0021 | >0.99 | >0.99 | 0.38 | >0.99 | >0.99 | >0.99 | <0.0001 | <0.0001 | 0.072 | <0.0001 | >0.99 |
| Sag ratio | 0.00011 | 0.00013 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | 0.99 | 0.022 | >0.99 | 0.10 | <0.0001 | 0.19 | 0.099 | >0.99 |
| First/last ISI ratio | <0.0001 | <0.0001 | 0.028 | 0.010 | >0.99 | 0.019 | >0.99 | >0.99 | 0.40 | >0.99 | 0.00036 | 0.0014 | 0.00031 | <0.0001 | >0.99 |
| Spike height (mV) | 0.036 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 |
| Spike width (ms) | <0.0001 | <0.0001 | <0.0001 | 0.0016 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | 0.0086 | <0.0001 | 0.0041 | <0.0001 | >0.99 |
| Pairwise comparisonsc |
|||||||||||||||
| Npas1+-Lhx6+ vs Npy2r+ | Npas1+-Lhx6+ vs PV+ | Npas1+-Lhx6+ vs PV+-Lhx6+ | Npas1+-Lhx6+ vs PV+-Npy2r+ | Npr3+ vs Npy2r+ | Npr3+ vs PV+ | Npr3+ vs PV+-Lhx6+ | Npr3+ vs PV+-Npy2r+ | Npy2r+ vs PV+ | Npy2r+ vs PV+-Lhx6+ | Npy2r+ vs PV+-Npy2r+ | PV+ vs PV+-Lhx6+ | PV+ vs PV+-Npy2r+ | PV+-Lhx6+ vs PV+-Npy2r+ | ||
| Spontaneous activity (Hz) | >0.99 | <0.0001 | 0.081 | 0.00050 | >0.99 | <0.0001 | 0.16 | 0.0050 | 0.029 | >0.99 | 0.26 | >0.99 | >0.99 | >0.99 | |
| CVISI | >0.99 | 0.0043 | >0.99 | 0.020 | >0.99 | 0.98 | >0.99 | 0.68 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | |
| Max rate (Hz) | 0.053 | <0.0001 | 0.11 | <0.0001 | 0.067 | <0.0001 | 0.13 | 0.00059 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | |
| Max current (pA) | 0.095 | 0.0018 | >0.99 | 0.050 | 0.038 | 0.0011 | >0.99 | 0.022 | >0.99 | >0.99 | >0.99 | 0.30 | >0.99 | >0.99 | |
| Membrane potential (mV) | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | |
| Trough potential (mV) | <0.0001 | <0.0001 | >0.99 | 0.00026 | 0.0015 | 0.0067 | >0.99 | 0.016 | >0.99 | 0.21 | >0.99 | >0.99 | >0.99 | >0.99 | |
| Sag ratio | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | |
| First/last ISI ratio | >0.99 | >0.99 | >0.99 | 0.28 | 0.64 | >0.99 | 0.79 | 0.090 | >0.99 | >0.99 | >0.99 | >0.99 | 0.28 | >0.99 | |
| Spike height (mV) | 0.47 | >0.99 | >0.99 | >0.99 | 0.33 | 0.75 | >0.99 | >0.99 | >0.99 | 0.30 | >0.99 | 0.61 | >0.99 | >0.99 | |
| Spike width (ms) | 0.0043 | <0.0001 | 0.0023 | <0.0001 | 0.27 | 0.0091 | 0.21 | 0.00038 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | >0.99 | |
aTwo-tailed p values are listed.
bKruskal–Wallis tests were performed to compare each characteristic across all groups.
cDunn tests were performed to compare each pair of neuron types.
The bolded p values are significant when using a Bonferroni correction at a significance level of 0.05, adjusting for the fact that 10 characteristics were considered.
To more effectively visualize how Npr3+ neurons, Kcng4+ neurons, Npy2r+ neurons, and PV+-Npy2r+ neurons co-cluster with other known GPe neuron types, we performed principal component analysis (PCA) and a variety of clustering approaches on the previously published datasets (Abecassis et al., 2020) along with our newly collected dataset. Hierarchical clustering yielded two clusters that are largely consistent with the PV+ class and Npas1+ class. As expected, Npr3+ neurons co-clustered with the Npas1+ neuron class, whereas Kcng4+ neurons and PV+-Npy2r+ neurons co-clustered with the PV+ neuron class. As there was no a priori information on whether PV+ neurons can be subdivided based on their electrophysiological attributes, we clustered all identified PV+ neurons (n = 100 neurons) separate from the rest of the dataset (Fig. 4a). t-SNE plots were used for visualizing high-dimensional data in a low-dimensional space. The relative distances between each marker represent similarities in the electrophysiological attributes. This analysis agreed with the PCA, in that two main clusters can be identified (Fig. 4b). The PCA shows that 45% of the total variance is explained by the first component and only 16% by the second component. The PCA biplot also shows the relative variance of each variable and their contribution to each principal component (Fig. 4c). Maximal firing rate, maximal current, spike width, and trough potential made the most contributions to the first principal component. In contrast, the first action potential height and membrane potential contributed the least.
Figure 4.
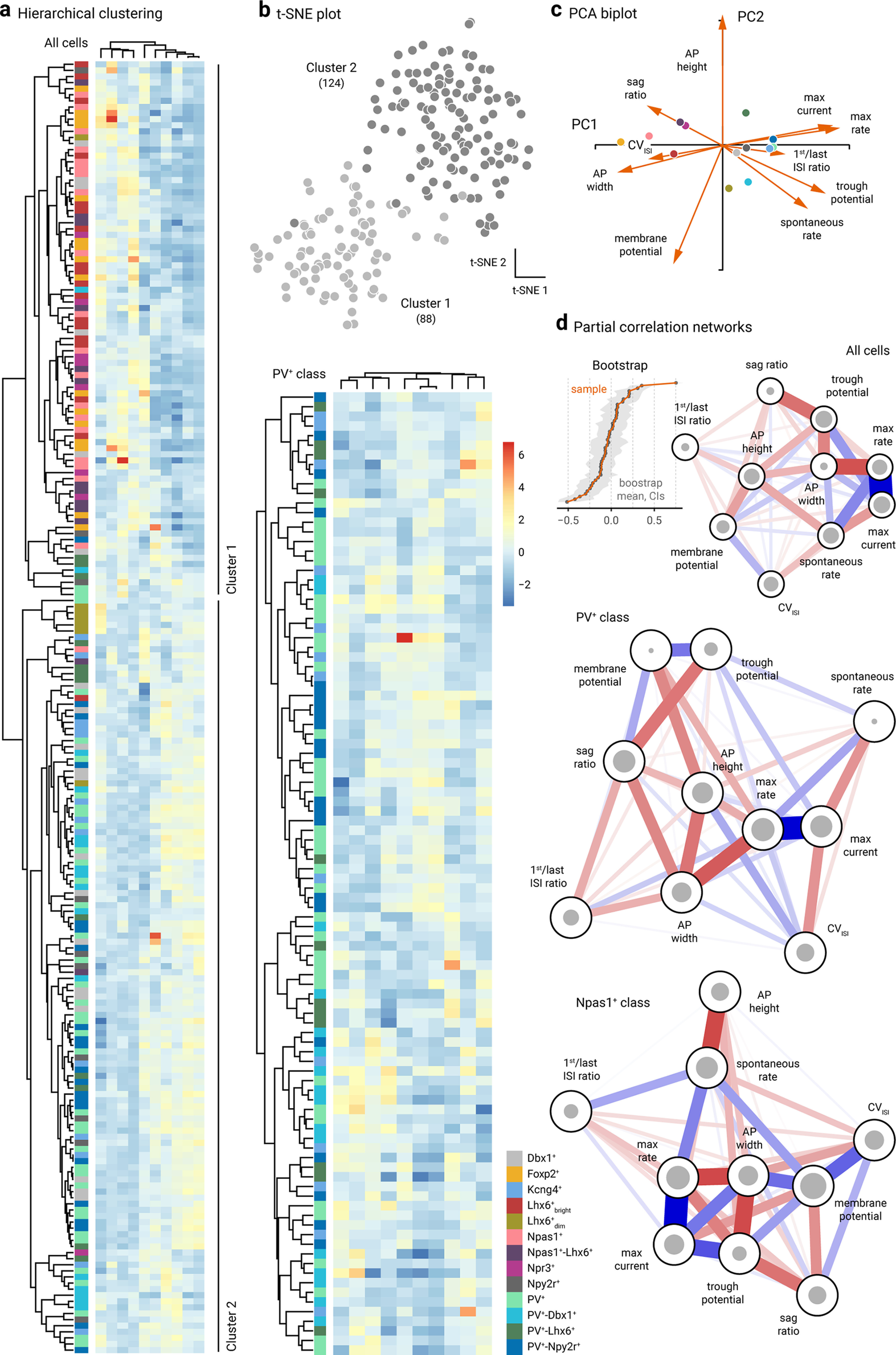
Clustering and network analysis of electrophysiological attributes. a, Left, Heatmap representation of electrical signatures of genetically-identified GPe neuron subtypes. Dendrograms represent the order and distances of neuron clusters and their electrical characteristics. A total of 212 neurons (n = Dbx1+: 20, Foxp2+: 16, Kcng4+: 13, Lhx6+bright: 18, Lhx6+dim: 7, PV+: 38, PV+-Dbx1+: 16, PV+-Lhx6+: 12, PV+-Npy2r+: 21, Npas1+: 19, Npas1+-Lhx6+: 13, Npr3+: 8, Npy2r+: 10) were included in this analysis. Neurons with incomplete data were excluded from the analysis. Two main clusters were identified: Cluster 1 contains 87 cells; Cluster 2 contains 125 cells. Column labels (left to right): membrane potential, CVISI, sag ratio, spike width, spike height, first/last ISI ratio, basal activity, trough potential, max rate, and max current. Right, Genetically-defined PV+ neurons are re-clustered. Scale bar applies to both panels. Purple-red-yellow represents Npas1+ neurons. Blue-green represents for PV+ neurons. Gray represents mixed neuron types (i.e., Dbx1+ and Npy2r+). Column labels (left to right): membrane potential, spike width, basal activity, trough potential, first/last ISI ratio, max rate, max current, sag ratio, CVISI, and spike height. b, t-SNE analysis of the same dataset as in a yielded two distinct clusters: Cluster 1 (light gray) contains 88 cells; Cluster 2 (dark gray) contains 124 cells. The membership assignment in Clusters 1 and 2 for each neuron subtypes is as follows: Foxp2+ (100.0%, 0.0%), Npas1+ (95.2%, 4.8%), Lhx6+bright (94.4%, 5.6%), Npr3+ (87.5%, 12.5%), Dbx1+ (30.0%, 70.0%), Npy2r+ (30.0%, 70.0%), PV+-Lhx6+ (25.0%, 75.0%), Lhx6+dim (14.3%, 85.7%), PV+-Dbx1+ (12.5%, 87.5%), PV+ (5.3%, 94.7%), PV+-Npy2r+ (4.8%, 95.2%), and Kcng4+ (0.0%, 100.0%). c, Principal component biplot showing the relative contributions of each electrophysiological attribute to PC1 and PC2. Markers represent the centroids for each cell type. The length of each vector is proportional to its variance with the cosine of the angle made by the vector with each axis indicating its contribution to that principal component. Highly correlated variables have similar directions; uncorrelated variables are perpendicular to each other. AP, action potential. d, Partial correlation networks with a spring layout from all cells (top), PV+ neurons (middle), and Npas1+ neurons (bottom). Inset, Bootstrap of edge stability. The size of the gray circles indicates the connectivity of nodes to each other as measured by closeness centrality index. Edge thickness indicates the strength of correlation between nodes. Colors represent the polarity of correlations. Blue represents positive correlations. Red represents negative correlations.
To better understand the relationship between individual attributes and how they differed between the two neuron classes, we performed network analysis with the dataset. Subtle differences between the PV+ neuron and Npas1+ neuron networks were found (Fig. 4d). A correlation between maximal firing rate, maximal current, and action potential width is consistent between the two networks and is conceivable as a function of congruent expression of voltage-gated sodium and potassium channels in these neurons (Hernández et al., 2015).
Markerless tracking effectively extracts behavioral dynamics
To investigate the roles of specific GPe populations in motor control, we analyzed the behavior of freely moving mice in an open field. We accomplished this by establishing a DeepLabCut (Mathis et al., 2018; Nath et al., 2019) model to track body kinematics (Fig. 5a). The model was trained until the loss reached a plateau. This trained network had a test error of 0.19 cm and a training error of 0.04 cm. This error was negligible relative to the mouse size: the median mouse width and length were 2.6 ± 0.28 and 6.0 ± 0.8 cm, respectively. Speeds computed from the tracking data generated from DeepLabCut and Ethovision were tightly correlated, confirming the validity of our model (Fig. 5c).
To translate movements into semantically meaningful behavioral features, we generalized four types of behavior in an open field: locomotion, fine movement, motionless, and rearing (Fig. 5a). The first three behaviors were classified by setting speed thresholds (see Materials and Methods). As rearing consists of complex movement sequences, we used SimBA, a machine learning tool, to generate a predictive classifier (Nilsson et al., 2020). This classifier has an F1 score = 0.71, precision = 0.68, and recall = 0.74 (Fig. 5c). Using this machine learning pipeline, we were able to effectively quantify behavioral events of mice in the open field in much greater detail and detect more complex, yet subtle, changes in body kinematics than before (Movie 1). We quantified the four different behaviors in PV-Cre and Npas1-Cre mice (n = 28 mice). At the population level, time spent in each category was: 55.8% in locomotion, 25.2% in fine movement, 10.0% in rearing, and 8.4% in motionless (Fig. 5b).
Movie 1.
Machine learning approaches capture mouse behavior motifs in an open field. Representative movie of a mouse in an open field arena. Current behavior of the mouse is displayed. Event timers track the cumulative duration of locomotion, rearing, fine movement, and motionless events. Video is shown at 0.5× speed.
Our previous studies established that PV+ neurons and Npas1+ neurons play causal but opposite roles in regulating locomotion—PV+ neurons are movement-promoting and Npas1+ neurons are movement-suppressing (Glajch et al., 2016; Pamukcu et al., 2020). Consistent with our previous observations, our analysis here showed that mice exhibited more mobile behavior (i.e., locomotion) when PV+ neurons were optogenetically stimulated and more immobile behavior (i.e., fine movement and motionless) when Npas1+ neurons were optogenetically stimulated. On the other hand, rearing was not consistently altered by optogenetic perturbations of either PV+ neurons or Npas1+ neurons (Fig. 5d). To monitor the orientation and posture of the mice, we measured the body angle and length-width ratio, respectively (Fig. 5e). A correlation matrix was constructed to visualize the relationships between different movement metrics (Fig. 5f). Spearman rank correlation tests between pairs of variables showed that 10 of the 21 pairs were correlated after the Bonferroni correction: locomotion was correlated with fine movement, rearing, body ratio, and speed; fine movement was correlated with motionless, body ratio, and speed; motionless was correlated with body ratio and speed; and body ratio was correlated with speed. In particular, the average event counts of fine movement and motionless were positively correlated with each other. Locomotion was associated with a high length-width ratio and a high speed, indicating that the mouse assumed an elongated form during locomotion. Fine movement and motionless were associated with a low length-width ratio and a low speed, which is indicative of a hunched and retracted posture. Similarly, body angle correlated with moment-to-moment changes in speed and body ratio (Fig. 5e). Collectively, these data provide a proof-of-concept that our analysis pipeline can be used effectively to extract body kinematics in mice.
Optogenetic manipulations induce cell-specific motor patterns
In addition to speed, body angle, and length-width ratio, we extracted 22 additional parameters to thoroughly capture motor pattern changes induced by optogenetic manipulations. As a large number of parameters (a total of 25), including movement sequences, were tracked, we examined the relationships between the measured parameters by constructing a correlation matrix for both PV-Cre and Npas1-Cre mice that were used in this study (n = 28). We found strong correlations between a large number of variable pairs (163 from 253), assessed via Spearman rank correlation tests after the Bonferroni correction. As mice have low incidences of transitioning between rearing and motionless, rearing-motionless and motionless-rearing switch frequencies were excluded from this analysis. At this point, it is unclear whether that was hardwired in the motor program or whether it was constrained by body mechanics. Although stimulation induced drastic changes in movements, the relationships between movement metrics remained unchanged. As such, the data from pre and light periods were combined. A condensed version of the correlation matrix that contains the most relevant metrics is presented in Figure 6a. For example, percent time spent on locomotion positively correlated (blue) with speed but negatively correlated (brown) with percent time spent on fine movement and motionless. To effectively visualize the behavioral responses in mice when PV+ neurons and Npas1+ neurons were optogenetically stimulated, we performed a PCA. As shown in Figure 6a, mice showed very similar behavior during the pre period. In contrast, they showed divergent responses when PV+ neurons and Npas1+ neurons were optogenetically stimulated during the light period.
Figure 6.
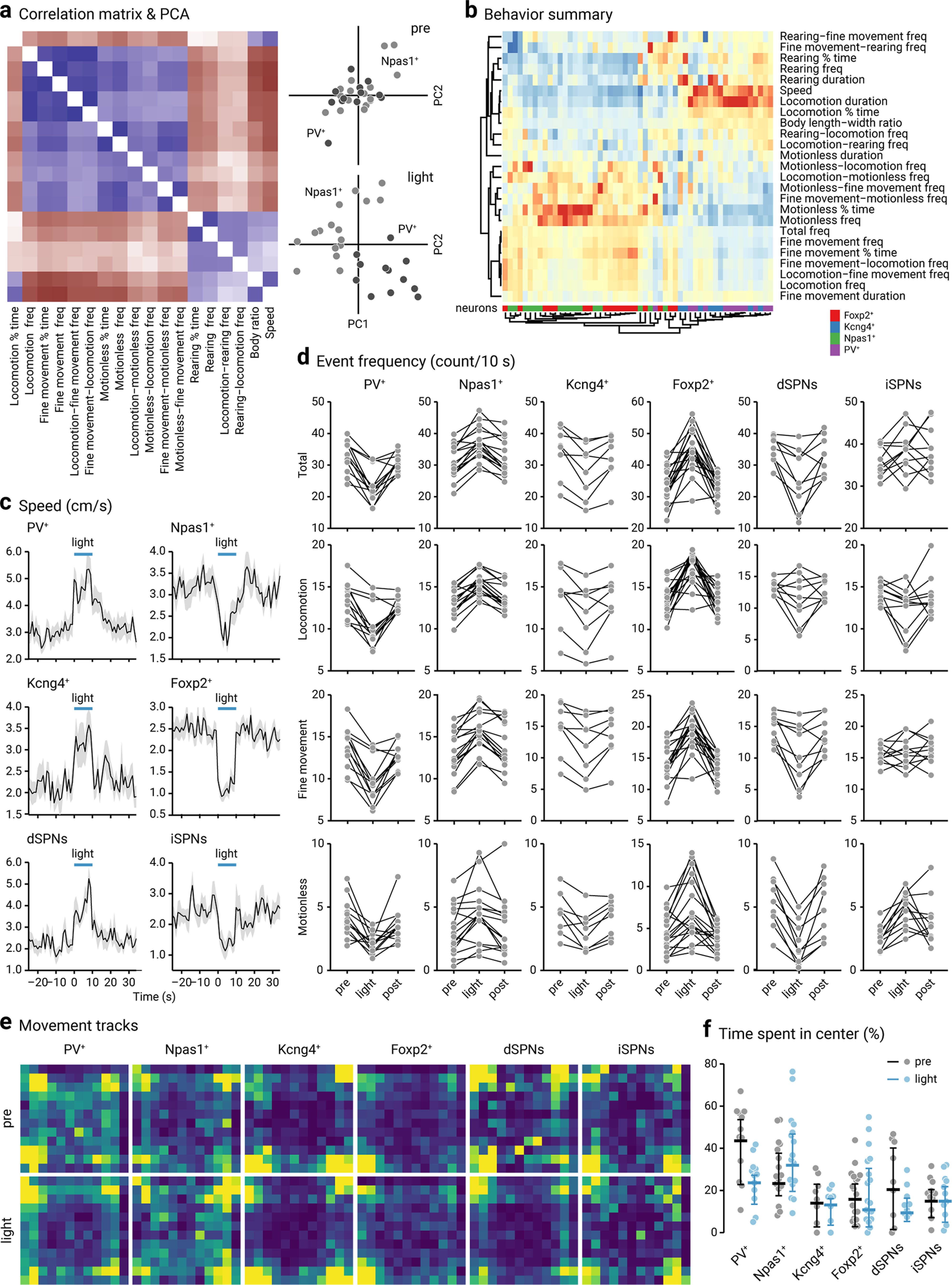
Motor patterns of mice with cell-specific optogenetic stimulation. a, Left, A correlation matrix constructed using data from PV-Cre (n = 12) and Npas1-Cre (n = 16) mice transduced with ChR2-eYFP. Eighteen parameters were included in this matrix. Blue represents positive correlations. Brown represents negative correlations. Right, Principal component analysis (PCA) plots showing the distributions of PV-Cre (dark gray, n = 12) and Npas1-Cre (light gray, n = 16) mice (transduced with ChR2-eYFP) in pre period (top) and light period (bottom). Each marker represents a mouse. Twenty-three movement metrics were used in this analysis. b, A heatmap summarizing motor responses of mice to optogenetic stimulation of genetically-defined neurons in the GPe. Twenty-five movement metrics were measured to fully capture the behavioral structures. Each of the 25 rows represents the fold change of movement metrics. Warm colors (red) represent positive changes. Cool colors (blue) represent negative changes. Rows and columns were sorted using hierarchical clustering. Dendrograms are divided into two main arms: metrics on the upper arm are negatively correlated with “total frequency,” whereas metrics on the lower arm are positively correlated with “total frequency.” Each column is a mouse; 54 mice were used in this analysis. The neurons of interest are PV+ (purple), Npas1+ (green), Kcng4+ (blue), and Foxp2+ (red) neurons. c, Plots showing the speed of mice with optogenetic stimulation in genetically-defined neurons in the GPe. Blue horizontal lines indicate the timing of light delivery. Thick lines indicate average. Gray areas represent SEM. dSPNs, direct-pathway striatal projection neurons; iSPNs, indirect-pathway striatal projection neurons. d, Slope graphs showing event frequency (counts per 10 s) for total, locomotion, fine movement, and motionless in mice and the effect with optogenetic stimulation of selective neuron types. Each connected line represents a mouse: PV+ neurons (n = 12), Npas1+ neurons (n = 16), Kcng4+ neurons (n = 8), Foxp2+ neurons (n = 18), dSPNs (n = 9), and iSPNs (n = 11). e, Heatmaps showing the relative cumulative time that mice spent in each position following optogenetic stimulation of genetically-defined GPe neurons. Yellow represents more time spent. Blue represents less time spent. Each heatmap was scaled to min and max and does not represent the net locomotor activity of mice. f, A univariate scatter plot summarizing the amount of time mice spent in the center of the area during pre (gray) and light period (blue). Thick lines indicate the medians. Whiskers represent interquartile ranges. PV+ neurons (n = 12), Npas1+ neurons (n = 16), Kcng4+ neurons (n = 8), Foxp2+ neurons (n = 18), dSPNs (n = 9), and iSPNs (n = 11).
Figure 6b is a heatmap representation of changes in movement metrics upon optogenetic stimulation of genetically-defined GPe neurons. Mice transduced with ChR2-eYFP in Foxp2+ neurons, Kcng4+ neurons, Npas1+ neurons, and PV+ neurons were included in this analysis. Hierarchical clustering revealed coordinated changes in behavioral dynamics across these test subjects. In particular, the stimulation of Foxp2+ neurons and Kcng4+ neurons led to changes that were consistent with those produced by the stimulation of Npas1+ neurons and PV+ neurons, respectively. Unlike the ethograms in Figure 5d that are purely event-counting across time, the heatmap in Figure 6b shows the fold changes of all 25 movement metrics in response to optogenetic stimulation. The most striking effects were the changes in speed, locomotion duration, percent time motionless, and motionless frequency. The raw data for speed is shown in Figure 6c. To contextualize the magnitude of the observed effect, direct- and indirect-pathway striatal projection neurons (dSPNs and iSPNs, respectively) were included for comparison. Figure 6d shows the changes in total, locomotion, fine movement, and motionless frequencies across individual mice. The subtle differences between mouse lines made it difficult to make arguments about quantitative differences. With this caveat in mind, it is nevertheless surprising to see that optogenetic stimulation of Kcng4+ neurons produced statistically indistinguishable effects on locomotor speed as PV+ neurons. This result was not expected as Kcng4+ neurons only account for ∼10% of the entirety of the GPe. Similarly, optogenetic stimulation of Foxp2+ neurons produced near-identical effects on locomotor speed as Npas1+ neurons (Table 4), while they only account for ∼25% of the entirety of the GPe (Fig. 1d).
Table 4.
Summary of optogenetic effects on behavior metrics
| PV+ neurons, ChR2 (n = 12) Fold change | pc | Npas1+ neurons, ChR2 (n = 16) Fold change | p | Kcng4+ neurons, ChR2 (n = 8) Fold change | p | Foxp2+ neurons, ChR2 (n = 18) Fold change | p | STN neurons, ACR2 (n = 10) Fold change | p | dSPNs, ChR2 (n = 9) Fold change | p | iSPNs, ChR2 (n = 11) Fold change | p | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Speed (cm/s) | 1.53 ± 0.31 | 0.00098 | 0.81 ± 0.09 | 0.000031 | 1.52 ± 0.33 | 0.0078 | 0.36 ± 0.12 | 0.0000076 | 1.68 ± 0.15 | 0.0020 | 1.9 ± 0.25 | 0.0039 | 0.52 ± 0.09 | 0.00098 |
| Body length-width ratio | 1.1 ± 0.05 | 0.00049 | 1.0 ± 0.05 | 0.00015 | 1.16 ± 0.08 | 0.023 | 0.88 ± 0.06 | 0.000023 | 1.19 ± 0.03 | 0.0020 | 1.14 ± 0.11 | 0.0039 | 0.97 ± 0.04 | 0.083 |
| Total frequencya | 0.71 ± 0.12 | 0.00049 | 1.08 ± 0.08 | 0.000061 | 0.79 ± 0.06 | 0.0078 | 1.36 ± 0.17 | 0.000015 | 0.68 ± 0.06 | 0.0020 | 0.76 ± 0.16 | 0.0039 | 1.12 ± 0.09 | 0.31 |
| Locomotion frequency | 0.78 ± 0.12 | 0.00049 | 1.1 ± 0.04 | 0.000092 | 0.82 ± 0.1 | 0.023 | 1.32 ± 0.18 | 0.000076 | 0.71 ± 0.05 | 0.0020 | 0.91 ± 0.18 | 0.20 | 0.9 ± 0.04 | 0.0098 |
| Fine movement frequency | 0.69 ± 0.12 | 0.00049 | 1.06 ± 0.1 | 0.000061 | 0.77 ± 0.13 | 0.0078 | 1.35 ± 0.21 | 0.000015 | 0.63 ± 0.08 | 0.0020 | 0.82 ± 0.14 | 0.0039 | 1.04 ± 0.08 | 0.81 |
| Rearing frequency | 0.91 ± 0.2 | 0.15 | 0.87 ± 0.18 | 0.026 | 0.98 ± 0.13 | 0.56 | 0.68 ± 0.31 | 0.00092 | 1.56 ± 0.33 | 0.0039 | 0.97 ± 0.32 | 0.97 | 0.29 ± 0.11 | 0.00098 |
| Motionless frequency | 0.5 ± 0.18 | 0.0015 | 1.03 ± 0.46 | 0.0011 | 0.67 ± 0.14 | 0.016 | 1.77 ± 0.34 | 0.0019 | 0.51 ± 0.13 | 0.0059 | 0.33 ± 0.14 | 0.0039 | 1.88 ± 0.34 | 0.00098 |
| Locomotion % time | 1.24 ± 0.09 | 0.0024 | 0.99 ± 0.07 | 0.0013 | 1.11 ± 0.04 | 0.0078 | 0.86 ± 0.06 | 0.0023 | 1.18 ± 0.13 | 0.0059 | 1.37 ± 0.12 | 0.0039 | 0.66 ± 0.1 | 0.0029 |
| Fine movement % time | 0.66 ± 0.14 | 0.00049 | 1.09 ± 0.14 | 0.000031 | 0.74 ± 0.16 | 0.0078 | 1.51 ± 0.32 | 0.000038 | 0.53 ± 0.12 | 0.0020 | 0.79 ± 0.13 | 0.0039 | 1.03 ± 0.08 | 0.64 |
| Rearing % time | 1.02 ± 0.25 | 0.91 | 0.82 ± 0.16 | 0.013 | 1.21 ± 0.14 | 0.023 | 0.54 ± 0.25 | 0.0010 | 1.42 ± 0.25 | 0.020 | 1.11 ± 0.57 | 0.65 | 0.39 ± 0.18 | 0.00098 |
| Motionless % time | 0.41 ± 0.15 | 0.0024 | 1.08 ± 0.49 | 0.0063 | 0.63 ± 0.2 | 0.0078 | 1.43 ± 0.53 | 0.060 | 0.5 ± 0.19 | 0.020 | 0.18 ± 0.1 | 0.0039 | 2.84 ± 0.34 | 0.00098 |
| Locomotion durationb | 1.83 ± 0.36 | 0.00049 | 0.91 ± 0.05 | 0.000031 | 1.38 ± 0.17 | 0.0078 | 0.57 ± 0.08 | 0.000015 | 1.58 ± 0.21 | 0.0020 | 1.7 ± 0.41 | 0.0039 | 0.95 ± 0.23 | 0.21 |
| Fine movement duration | 0.96 ± 0.03 | 0.016 | 1.01 ± 0.06 | 0.011 | 0.94 ± 0.04 | 0.039 | 1.16 ± 0.1 | 0.0010 | 0.85 ± 0.08 | 0.0098 | 0.93 ± 0.07 | 0.027 | 0.94 ± 0.05 | 0.019 |
| Rearing duration | 1.17 ± 0.13 | 0.11 | 0.99 ± 0.11 | 0.074 | 1.34 ± 0.49 | 0.31 | 0.7 ± 0.12 | 0.0066 | 0.92 ± 0.12 | 0.16 | 0.84 ± 0.29 | 0.82 | 1.28 ± 0.56 | 0.43 |
| Motionless duration | 0.78 ± 0.1 | 0.0093 | 0.95 ± 0.09 | 0.86 | 0.95 ± 0.13 | 0.38 | 0.88 ± 0.11 | 0.067 | 1.0 ± 0.28 | 0.63 | 0.61 ± 0.08 | 0.0039 | 1.67 ± 0.32 | 0.0020 |
| Locomotion-fine movement frequency | 0.77 ± 0.14 | 0.00049 | 1.08 ± 0.08 | 0.000061 | 0.79 ± 0.17 | 0.039 | 1.32 ± 0.26 | 0.00033 | 0.62 ± 0.04 | 0.0020 | 1.01 ± 0.24 | 0.50 | 0.72 ± 0.13 | 0.0020 |
| Locomotion-rearing frequency | 1.02 ± 0.13 | 0.93 | 1.0 ± 0.18 | 0.75 | 0.87 ± 0.11 | 0.054 | 0.76 ± 0.11 | 0.083 | 1.43 ± 0.07 | 0.0020 | 1.14 ± 0.13 | 0.31 | 0.57 ± 0.29 | 0.031 |
| Locomotion-motionless frequency | 0.7 ± 0.21 | 0.034 | 1.06 ± 0.31 | 0.021 | 0.88 ± 0.25 | 0.64 | 1.37 ± 0.35 | 0.0019 | 0.73 ± 0.21 | 0.29 | 0.77 ± 0.06 | 0.0039 | 1.38 ± 0.35 | 0.0088 |
| Fine movement-locomotion frequency | 0.77 ± 0.11 | 0.00049 | 1.06 ± 0.11 | 0.00015 | 0.79 ± 0.16 | 0.039 | 1.35 ± 0.24 | 0.00011 | 0.64 ± 0.08 | 0.0020 | 1.02 ± 0.3 | 0.48 | 0.8 ± 0.12 | 0.0020 |
| Fine movement-rearing frequency | 0.95 ± 0.08 | 0.12 | 1.0 ± 0.22 | 0.39 | 0.99 ± 0.08 | >0.99 | 0.96 ± 0.16 | 0.54 | 0.97 ± 0.2 | 0.47 | 0.86 ± 0.46 | 0.44 | 0.0 ± 0.0 | >0.99 |
| Fine movement-motionless frequency | 0.71 ± 0.15 | 0.034 | 1.06 ± 0.2 | 0.034 | 0.72 ± 0.12 | 0.078 | 1.31 ± 0.36 | 0.048 | 0.6 ± 0.11 | 0.063 | 0.45 ± 0.11 | 0.0039 | 1.56 ± 0.36 | 0.00098 |
| Rearing-locomotion frequency | 1.09 ± 0.16 | 0.83 | 1.03 ± 0.18 | 0.9 | 0.95 ± 0.16 | 0.74 | 0.81 ± 0.19 | 0.086 | 1.17 ± 0.06 | 0.027 | 1.07 ± 0.18 | 0.81 | 0.67 ± 0.17 | 0.0078 |
| Rearing-fine movement frequency | 1.0 ± 0.0 | 0.25 | 1.0 ± 0.15 | 0.57 | 0.95 ± 0.06 | 0.13 | 0.9 ± 0.13 | 0.49 | 0.78 ± 0.28 | 0.13 | 1.0 ± 0.14 | 0.63 | 1.0 ± 0.33 | >0.99 |
| Motionless-locomotion frequency | 0.71 ± 0.13 | 0.077 | 1.01 ± 0.33 | 0.0082 | 0.92 ± 0.17 | 0.74 | 1.31 ± 0.33 | 0.054 | 0.78 ± 0.13 | 0.027 | 0.71 ± 0.07 | 0.0078 | 1.42 ± 0.27 | 0.0059 |
| Motionless-fine movement frequency | 0.72 ± 0.12 | 0.027 | 0.96 ± 0.39 | 0.074 | 0.71 ± 0.09 | 0.023 | 1.36 ± 0.25 | 0.027 | 0.72 ± 0.31 | 0.29 | 0.43 ± 0.11 | 0.0039 | 1.67 ± 0.35 | 0.00098 |
aUnit for all frequencies is count/10 s.
bUnit for all durations is seconds.
cWilcoxon test; two-tailed exact p values are shown.
The bolded p values are significant when using a Bonferroni correction at a significance level of 0.05.
Given Kcng4+ neurons are ∼30% (28 ± 8%, n = 3 sections, 121 neurons) of PV+ neurons and Foxp2+ neurons are ∼60% (57 ± 4%, n = 6 sections, 566 neurons) of Npas1+ neurons, our results raised a question of whether PV+-Kcng4– neurons and Npas1+-Foxp2– neurons could be involved in nonmotor function. Close examination of the movement tracks revealed that optogenetic stimulation of PV+ neurons and Npas1+ neurons led to changes in the anxiety-like behavior, as measured by their center-edge preference in the open-field arena. As shown in Figure 6e, f, PV+ neuron stimulation decreased the time spent at the center, whereas Npas1+ neuron stimulation increased it (PV+ pre: 47.3 ± 10.3%, PV+ light: 27.4 ± 5.4%, n = 12, p = 0.0034; Npas1+ pre: 27.3 ± 10.4%, Npas1+ light: 35.7 ± 12.3%, n = 16, p = 0.0060). These effects were highly cell type-specific as they were not observed with the stimulation of Kcng4+ neurons, Foxp2+ neurons, dSPNs, or iSPNs and thus were dissociable from the locomotor effects (Kcng4+ pre: 17.7 ± 7.9%, Kcng4+ light: 16.9 ± 4.1%, n = 8, p = 0.38; Foxp2+ pre: 11.5 ± 10.7%, Foxp2+ light: 10.4 ± 8.7%, n = 18, p = 0.30; dSPN pre: 24.1 ± 19.0%, dSPN light: 13.1 ± 4.1%, n = 9, p = 0.10; iSPN pre: 18.6 ± 7.2%, iSPN light: 18.6 ± 9.6%, n = 11, p = 0.90).
Combinatorial movement metrics infers behavioral states
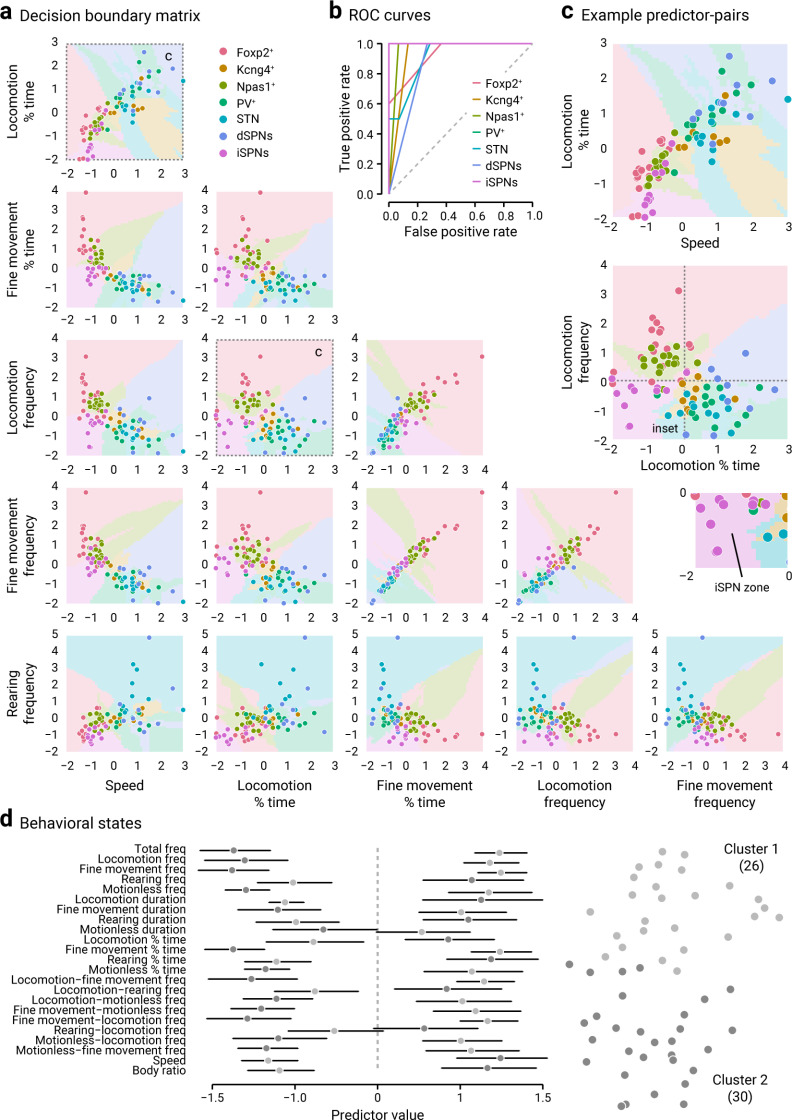
To visualize how each neuron subtype would be classified based on their motor effects upon optogenetic stimulation, we constructed a decision boundary matrix using a k-nearest neighbors classification algorithm, in which multiple movement metrics were considered (Fig. 7a). This contrasts with the analysis shown in Figure 6d, in which only a single movement metric was compared. As supported by the receiver operating characteristic curves in Figure 7b, each neuron type studied here gave rise to unique behavior structures. This analysis showed that a number of neuron types occupied a unique parameter space as delineated by the decision boundaries visualized (e.g., Fig. 7c). As shown in the relationship between locomotion frequency and the percent time in locomotion (Fig. 7c), PV+ neurons and Npas1+ neurons were on opposite ends of the spectrum. This suggests that PV+ neurons and Npas1+ neurons work against each other. On the other hand, PV+ neurons, Kcng4+ neurons, dSPNs, and STN neurons (transduced with GtACR2, an inhibitory opsin) were close to one another, suggesting that these neurons work either together or converge on a common downstream target to mediate the same action. This idea is supported by experimental evidence that PV+ neurons and Kcng4+ neurons send inhibitory signals to the SNr, which receive direct inhibitory inputs from dSPNs (Grofova, 1975; Araki et al., 1985; Bolam et al., 1993; Mink, 1996; Smith et al., 1998; Connelly et al., 2010; Cui et al., 2021). In contrast, iSPNs that are known to heavily target PV+ neurons (but not PV– neurons) (Yuan et al., 2017) are on the bottom left corner of the plot. This analysis reinforced the notion that the differences in the tracked motor behaviors were in fact cell-specific and not simply esoteric fluctuations.
Figure 7.
Optogenetic manipulations produced cell-specific motor patterns. a, A decision boundary matrix visualizing the relationships between movement metrics and how they would be classified if predicted through the current model. Boundaries between classes are visualized. All variables were scaled to have a mean of 0 and an SD of 1. Each circle represents a mouse. b, Receiver operating characteristic (ROC) curves for different cell types are shown. Classifiers that give curves closer to the top left corner indicate a better performance; the closer the curve comes to the diagonal of the ROC space, the less accurate the test. c, Magnified view of the relationships between two predictor pairs. Hash marks indicate the origin. Data associated with iSPNs are uniquely situated on the bottom left corner of the graph (see Inset). d, Left, Summary plot showing the mean and its 95% CI for each of 23 variables for each of the two clusters. All variables were scaled. Right, t-SNE visualization of clustering. Cluster 1 (light gray) has a size of 26, whereas Cluster 2 (dark gray) has a size of 30. Memberships for Cluster 1: PV+ pre = 41.7% (5/12); PV+ light = 75.0% (9/12); Npas1+ pre = 56.3% (9/16); Npas1+ light = 18.8% (3/16). Memberships for Cluster 2: PV+ pre = 58.3% (7/12); PV+ light = 25.0% (3/12); Npas1+ pre = 43.8% (7/16), Npas1+ light = 81.3% (13/16).
As noted earlier, as PV+ neurons and Npas1+ neurons produced diametric behavioral effects, we asked whether PV+ neurons and Npas1+ neurons could be working in concert to regulate the transitions between behavioral states in mice. Instead of simply providing granularity of behavioral dynamics, we sought to determine whether we can use the different movement metrics collectively to define behavioral states. To this end, we parsed out the data from both PV-Cre and Npas1-Cre mice during both pre and light periods and then performed a k-means clustering (with k = 2) that included all 25 movement metrics. In Figure 7d, the dimensionality of the data is reduced via t-SNE, with each mouse shaded according to which cluster it is assigned. As expected, the predicted cluster memberships for Clusters 1 and 2 were about evenly split in both PV-Cre and Npas1-Cre mice during the pre period (PV: 56.3%; Npas1: 41.7%). However, optogenetic stimulation shifted the membership distributions during the light period: PV-Cre mice were predominantly (75.0%) in Cluster 1, whereas Npas1-Cre were predominantly (81.3%) in Cluster 2.
The k-means clustering analysis thus provided critical insights. First, it reinforced the idea that optogenetic manipulations of PV+ neurons and Npas1+ neurons give rise to distinct physiological behaviors. Second, the analysis suggests that the combinations of movement metrics can be used to define two behavioral states and supports the notion that PV+ neurons and Npas1+ neurons act as opposing forces that tune mouse behavior.
Local collateral connectivity within the GPe is cell-specific
Based on the opposing roles of PV+ neurons and Npas1+ neurons, we hypothesized that they communicate with each other via inhibitory connections. Consistent with prior observations from single-cell labeling studies (Kita and Kitai, 1994; Nambu and Llinás, 1997; Bevan et al., 1998; Sato et al., 2000; Sadek et al., 2007; Fujiyama et al., 2016), biocytin-filled GPe neurons produced local collaterals that contain large varicosities that can be readily observed post hoc with confocal microscopy (Fig. 8a), although the postsynaptic partner cannot be easily identified with this approach.
Figure 8.
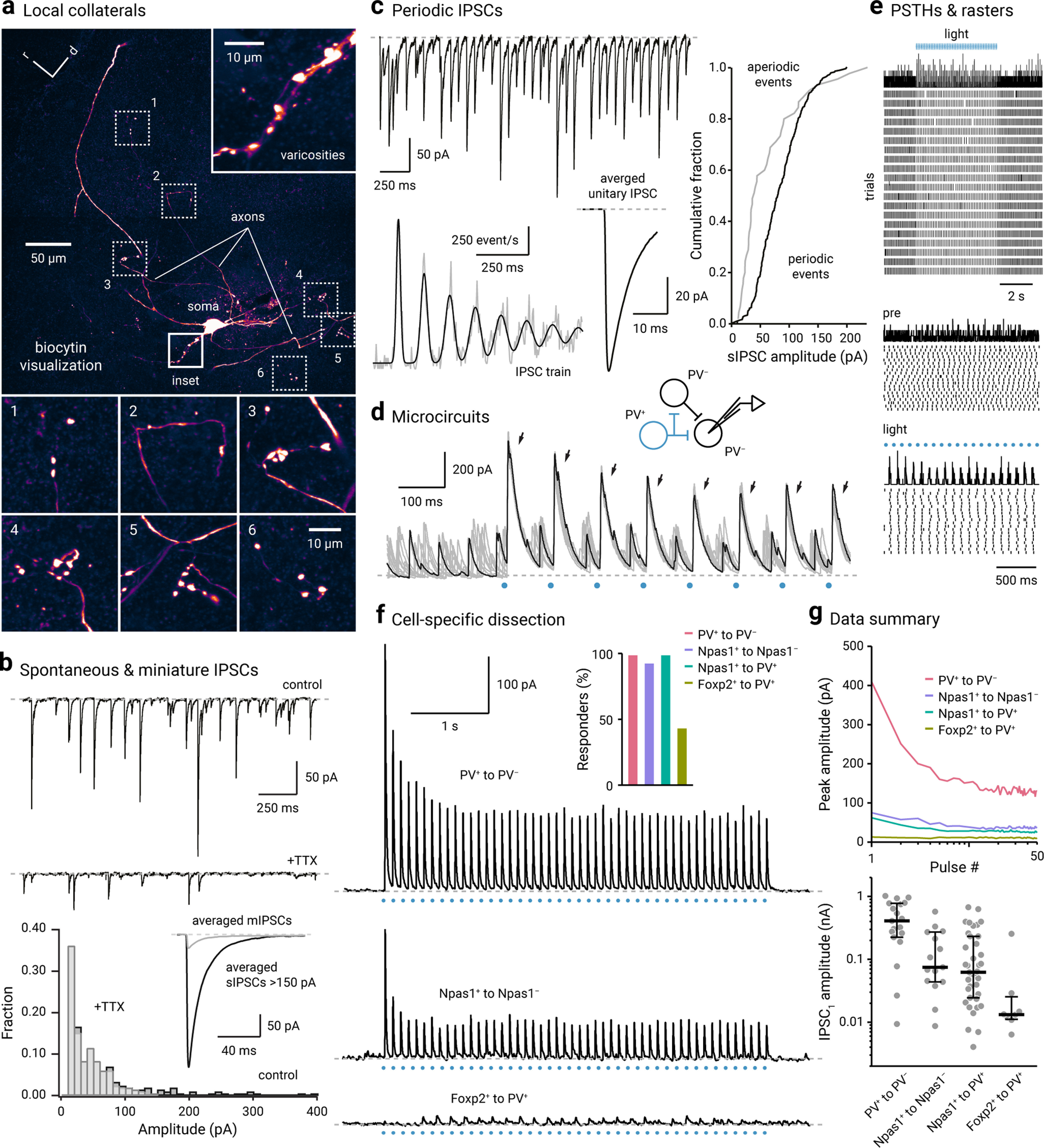
Local collateral connectivity within the GPe is cell-specific. a, An axon arborized locally in the vicinity of a biocytin-filled GPe neuron. Axons were identified as small-caliber processes that bear varicosities and contain no spines. Top right, Inset, A series of varicosities produced by the biocytin-filled neuron. Bottom, Six additional examples of varicosities and their parent axons are shown. b, Top, Inwardly directed spontaneous inhibitory postsynaptic currents (IPSCs) were readily observed in voltage-clamped GPe neurons ex vivo. Large, periodic events were blocked by the application of tetrodotoxin (TTX) (1 μm), a voltage-gated sodium channel blocker. Bottom, Histogram showing the IPSC amplitude distribution. Inset, Averaged miniature IPSCs (gray) and large (>150 pA) IPSCs (black) are shown. c, Top, A current trace showing periodic IPSCs recorded from a voltage-clamped GPe neuron. Bottom, An autocorrelation of the IPSC train from an example neuron with one unitary input, fitted with a sum of Gaussian components, indicating that the neuron received one periodic input in addition to aperiodic inputs (left). The waveform of the averaged unitary IPSC is shown (right). Right, A cumulative plot showing the difference in the amplitude distribution of periodic (black) and aperiodic events (gray). d, Spontaneous IPSCs and optogenetically evoked IPSCs (arrows) were observed from a voltage-clamped PV– neuron (Vhold = –30 mV). A low-chloride internal solution was used. Light stimuli (blue circle) were delivered asynchronously relative to the occurrence of spontaneous IPSCs. A proposed microcircuit is shown on the top. e, Top, Peristimulus time histogram (PSTH) and raster plot showing the occurrence of spontaneous IPSCs. Evoked IPSCs were excluded. Middle and Bottom, An expanded time scale showing the timing of spontaneous IPSCs was reset by evoked IPSCs. Periodicity is demonstrated by the evenly spaced peaks in the PSTH. No temporal structures in the spontaneous IPSCs were observed before light delivery. f, Local connectivity was dissected using optogenetics ex vivo. Example traces from PV+ to PV– (top), Npas1+ to Npas1– (middle), and Foxp2+ to PV+ (bottom) are shown. ChR2-eYFP negative neurons were targeted for recording. For positive identification of PV+ neurons, PV+ neurons were labeled with the PV-tdTomato allele. Inset, Four connection types were examined: PV+ to PV– (blush, n = 19), Npas1+ to Npas1– (purple, n = 16), Npas1+ to PV+ (green, n = 39), and Foxp2+ to PV+ (olive, n = 18). The proportion of neurons that responded are shown as bar graphs. Only a subset of recorded PV+ neurons (8 of 18, 44.4%) showed detectable Foxp2+ input. g, Top, Population data showing IPSC amplitude for four different connection types across 50 pulses: PV+ to PV– (blush, n = 19), Npas1+ to Npas1– (purple, n = 15), Npas1+ to PV+ (green, n = 39), and Foxp2+ to PV+ (olive, n = 8). Each line indicates median values. Bottom, Univariate scatter plot showing the first IPSC amplitudes for four different connection types. Thick lines indicate the medians. Whiskers represent interquartile ranges. Each circle represents a neuron.
The presence of local inhibitory networks was further supported by the presence of large, periodic inhibitory postsynaptic currents in voltage-clamp recordings with a high-chloride internal solution. As expected from the local rather than extrinsic (e.g., from the striatum) origin, these events are action potential-dependent, as demonstrated by their sensitivity to the application of tetrodotoxin (1 μm), a voltage-gated sodium channel blocker (Fig. 8b). To gain further insights into the local connectivity, spontaneous inhibitory postsynaptic currents (IPSCs) were recorded from PV+ neurons and Npas1+ neurons; properties of periodic and aperiodic IPSCs were extracted using our established analysis routine (Higgs and Wilson, 2016) (Fig. 8c). In agreement with the results in Figure 8b, the small-amplitude aperiodic events were likely of striatal origin, whereas the large-amplitude periodic events likely arose from local collateral inputs. Furthermore, this analysis revealed that PV+ neurons and Npas1+ neurons receive local inputs and display IPSCs with biophysical properties that were not different from each other (Table 5).
Table 5.
Properties of spontaneous IPSCs in PV+ neurons and Npas1+ neurons
| PV+ neurons Median ± MAD | na | Npas1+ neurons Median ± MAD | n | Mann–Whitneyb | ||
|---|---|---|---|---|---|---|
| All spontaneous IPSCs | Rate (Hz) | 12.89 ± 3.63 | 50 | 7.76 ± 2.90 | 47 | 0.025 |
| Amplitude (pA) | 71.00 ± 20.54 | 50 | 60.24 ± 13.11 | 47 | 0.037 | |
| Autocorrelation fit parameters | Aperiodic rate (Hz) | 8.01 ± 3.13 | 12 | 7.37 ± 1.34 | 10 | >0.99 |
| Presynaptic frequency (Hz) | 8.88 ± 2.03 | 13 | 9.75 ± 1.14 | 11 | 0.49 | |
| Periodic rate (Hz) | 6.69 ± 1.17 | 13 | 7.25 ± 1.50 | 11 | 0.56 | |
| Presynaptic CVISI | 0.13 ± 0.04 | 13 | 0.15 ± 0.06 | 11 | 0.28 | |
| Power parameter | 0.70 ± 0.23 | 13 | 0.72 ± 0.10 | 11 | 0.88 | |
| Unitary IPSCs | Amplitude (pA) | 133.64 ± 71.95 | 12 | 82.70 ± 40.25 | 10 | 0.20 |
| CV | 0.50 ± 0.16 | 12 | 0.56 ± 0.10 | 10 | 0.54 | |
| Amplitude with failures (pA) | 111.91 ± 68.80 | 12 | 72.86 ± 39.60 | 10 | 0.25 | |
| CV with failures | 0.80 ± 0.34 | 12 | 0.85 ± 0.11 | 10 | 0.82 | |
| Variance/mean with failures | 79.00 ± 11.15 | 12 | 34.36 ± 10.50 | 10 | 0.15 |
an equal to the number of neurons.
bTwo-tailed exact p values are listed. No results are significant after applying the Bonferroni correction at a significance level of 0.05.
While it is probable that PV+ neurons and Npas1+ neurons are reciprocally connected to each other via inhibitory connections, the precise local connectivity patterns between GPe neurons have not been examined previously. As termination sites produced by local axonal arborizations are sparse and spatially distributed within the GPe (Kita and Kitai, 1994; Nambu and Llinás, 1997; Bevan et al., 1998; Sato et al., 2000; Sadek et al., 2007; Bugaysen et al., 2013; Fujiyama et al., 2016), an optogenetic approach instead of paired recordings was used to more efficiently survey the local connectivity patterns. ChR2-eYFP was transduced into genetically-defined neurons in PV-Cre and Npas1-Cre mice, using a Cre-inducible approach. Transgene negative neurons were targeted for voltage-clamp recordings. As illustrated in Figure 8d, optogenetic stimulation of PV+ neurons induced antiphase timing of a spontaneous synaptic input, suggesting antiphase spiking of a putative PV– neuron. This is more clearly shown in the peristimulus time histogram in Figure 8e. These results highlight the potential of local collateral input to shape population activity across the GPe. Using similar approaches, we tested the connectivity between various neuron types (PV+ to PV–, Npas1+ to Npas1–, Npas1+ to PV+, and Foxp2+ to PV+) (Fig. 8f,g). To avoid confounds associated with photocurrents, ChR2-eYFP negative neurons were targeted for recording. We found that PV+ to PV– input was the strongest of all connection types examined (PV+ to PV–: 409.9 ± 233.0 pA, n = 19 neurons; Npas1+ to Npas1–: 75.1 ± 59.2 pA, n = 15 neurons; Npas1+ to PV+: 62.2 ± 44.0 pA, n = 39 neurons; Foxp2+ to PV+: 13.3 ± 2.0 pA, n = 8 neurons; PV+ to PV– vs Npas1+ to Npas1–, p = 0.0024; PV+ to PV– vs Npas1+ to PV+, p < 0.0001; PV+ to PV– vs Foxp2+ to PV+, p = 0.00023; Npas1+ to Npas1– vs Npas1+ to PV+, p = 0.53; Npas1+ to Npas1– vs Foxp2+ to PV+, p = 0.0053; Npas1+ to PV+ vs Foxp2+ to PV+, p = 0.0047; Table 6). This finding is consistent with the earlier observation that PV+ boutons are abundant and form basket-like aggregates on cell bodies of both PV+ and PV– neurons within the GPe (Kita, 1994). Although Npas1+ neurons did target PV+ (which are Npas1–) neurons, the strength was smaller than that of the PV+ to PV– connection. The asymmetry in the connectivity between PV+ neurons and Npas1+ neurons can be partially explained by their abundance. Additional factors, such as the number of release sites, quantal content, and postsynaptic receptor density, may also be at play.
Table 6.
Local collateral connectivity within the GPe
| Input | IPSC measure |
Statistical analysesa |
|||||
|---|---|---|---|---|---|---|---|
| Median ± MAD (pA) | Response rate (%) | nb | vs PV+ to PV– | vs Npas1+ to Npas1– | vs Npas1+ to PV+ | vs Foxp2+ to PV+ | |
| PV+ to PV– | 409.9 ± 233.0 | 100 | 19 | — | 0.0024 | <0.0001 | 0.00023 |
| Npas1+ to Npas1– | 75.1 ± 59.2 | 93.8 | 16 | 0.0024 | — | 0.53 | 0.0053 |
| Npas1+ to PV+ | 62.2 ± 44.0 | 100 | 39 | <0.0001 | 0.53 | — | 0.0047 |
| Foxp2+ to PV+ | 13.3 ± 2.0 | 44.4 | 18 | 0.00023 | 0.0053 | 0.0047 | — |
aMann–Whitney U test on peak amplitude; two-tailed exact p values are shown.
bSample size (n) included all cells with or without responses.
The bolded p values are significant when using a Bonferroni correction at a significance level of 0.05, adjusting for the fact that 6 comparisons were considered.
Only a subset of recorded PV+ neurons (8 of 18, 44.4%) showed detectable Foxp2+ input, and the strength of this connection was the weakest among all connection types examined (Foxp2+ to PV+ vs PV+ to PV–, p = 0.00023; Foxp2+ to PV+ vs Npas1+ to Npas1–, p = 0.0053; Foxp2+ to PV+ vs Npas1+ to PV+, p = 0.0047) (Table 6). The observation of this weak and likely biologically negligible connection is highly consistent with recent studies (Aristieta et al., 2020; Ketzef and Silberberg, 2020). As it was clear that the Npas1+ input targets PV+ neurons, these results collectively imply that PV+ neurons receive Npas1+ input largely from the Nkx2.1+ (also known as Npr3+) subpopulation, which do not express Foxp2 (Fig. 1c) (see also Abecassis et al., 2020).
Discussion
In this study, we determined that Npr3+ neurons shared the unique molecular and anatomical signature of Npas1+-Nkx2.1+ neurons. While Kcng4+ neurons share canonical properties with PV+ neurons, they were not involved in regulating anxiety-like behavior (as measured with center-edge preferences). By using machine learning and statistical approaches to delineate the functional properties of GPe neuron subtypes, we proposed that PV+ neurons and Npas1+ neurons work in concert to control the transitions between behavioral states in mice. This idea was further supported by the existence of local connectivity between them.
GPe neuron diversity
What is a cell type, and why is it an important question? Both the embryonic origins and the congruent expression of transcription factors govern cell specifications (Hu et al., 2017; Lim et al., 2018; Fishell and Kepecs, 2020). It is well established that the GPe primarily arises from the medial and lateral ganglionic eminence (Flandin et al., 2010; Nóbrega-Pereira et al., 2010; Dodson et al., 2015; Y. J. Chen et al., 2017). It is clear that Npas1+ neurons are derived from both progenitor pools; Foxp2+ neurons are from the lateral ganglionic eminence, whereas Npr3+ neurons are from the medial ganglionic eminence and maintain the expression of Nkx2.1 (Fig. 1c). Consistent with the single-cell transcriptomic data (Saunders et al., 2018), our current and past (Abecassis et al., 2020) data showed the molecular complexity of PV+ neurons (Fig. 9). This is in agreement with earlier studies that hinted at multiple medial ganglionic eminence progenitors giving rise to at least two populations of PV+ neurons (Silberberg et al., 2016). In fact, heterogeneity among PV+ neurons can be a general feature across brain areas (Dehorter et al., 2015; Garas et al., 2016; Tasic et al., 2018; Hodge et al., 2019; Huang and Paul, 2019; Mahadevan et al., 2020). It has been shown that there is sonic hedgehog expression in the preoptic area and in the nascent GPe (Flandin et al., 2010; Sousa and Fishell, 2010). In agreement with this, earlier studies on Dbx1+ neurons showed that preoptic area neurons make a small contribution to the cellular makeup of the GPe (Gelman et al., 2011; Abecassis et al., 2020). This inference was confirmed with our recent, progenitor-specific fate-map study (Poulin et al., 2020).
Figure 9.
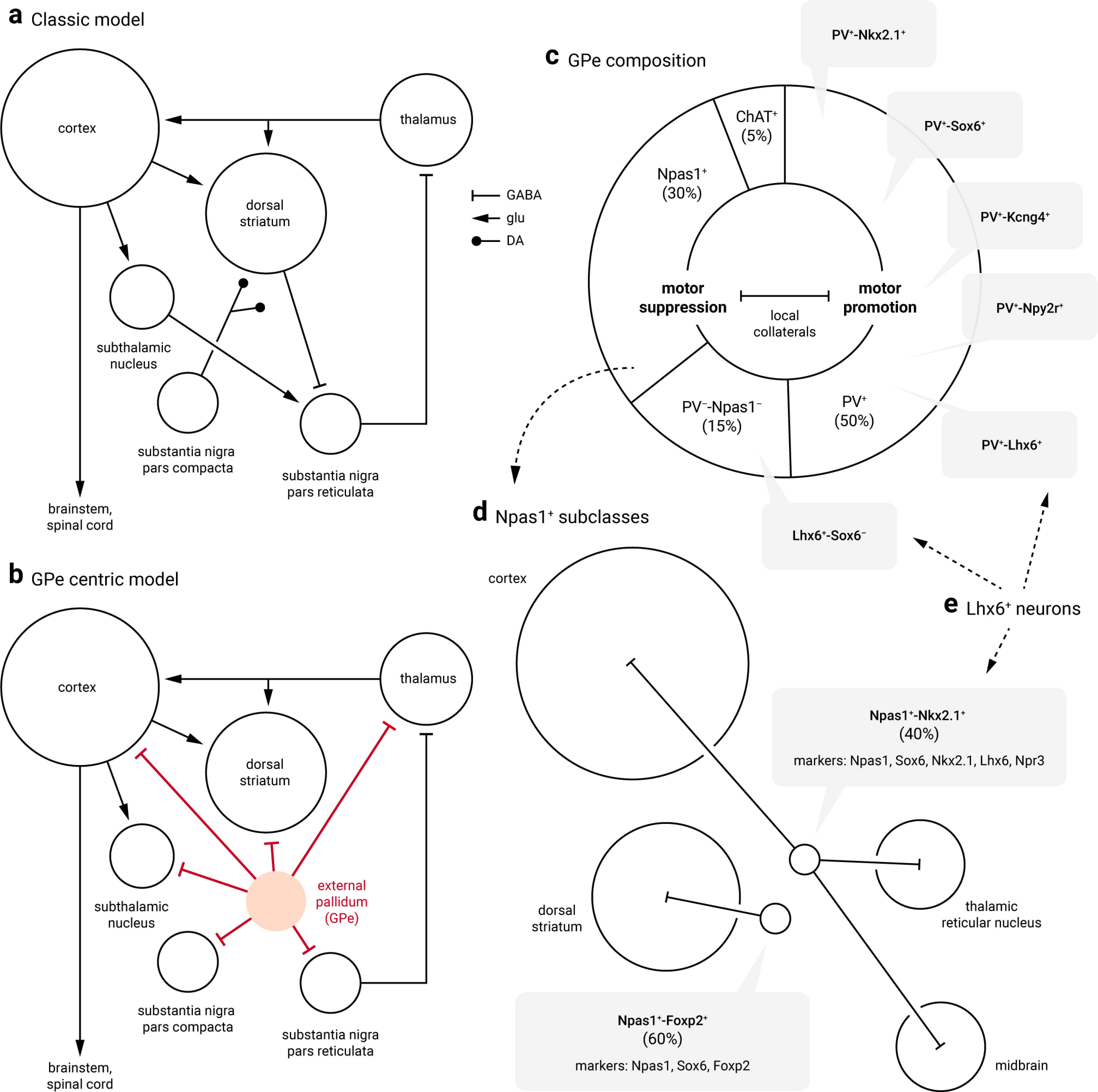
Cellular and circuit substrates for motor regulation by the GPe. a, A simplified circuit diagram represents the key nodes within the basal ganglia. The dStr and STN are the two input stations that receive excitatory cortical inputs; the SNr is the output station that provides inhibitory projection to the thalamus. The entire basal ganglia circuit, especially the dStr, is under the neuromodulatory control of the SNc. Size of the target areas (circles) is an artistic rendering based on the volume of the brain areas. The circuit role of the GPe is underappreciated. The GPe was traditionally viewed as a “black box,” which relays information from the dStr to the STN. b, A modified circuit model highlighting the central role of the GPe within the basal ganglia. The GPe sends inhibitory projections to all key structures within the circuit. c, Pie chart summarizing the neuronal composition of the mouse GPe. The area of the sectors represents the approximate size of each neuron class. PV+ neurons (which constitute 50% of the GPe) are heterogeneous. Nkx2.1, Sox6, Kcng4, Npy2r, and Lhx6 are coexpressed in PV+ neurons to a varying extent. How they intersect with each other remains to be determined. Previous transcriptomic data point to the idea that Kcng4+ neurons represent a unique subclass of PV+ neurons in the GPe. By projecting to the STN and SNr, PV+ neurons assume the canonical role of the GPe. Npas1+ neurons are 30% of the GPe; they can be subdivided into two subclasses (see d). Consistent with the distinct electrophysiological properties, PV+ neurons and Npas1+ neurons have unique behavioral roles: PV+ neurons are motor-promoting, whereas Npas1+ neurons are motor-inhibiting. PV+ neuron and Npas1+ neuron classes jointly tune motor behavior through a push-pull mechanism. Local inhibitory connections between these neuron classes serve as key cellular substrates that underlie this process. ChAT+ neurons are ∼5% of the total GPe neuron population and show no overlap with other known classes of GPe neurons. They project broadly to cortical areas. PV−-Npas1− neurons amount to ∼15% of the total GPe and are the least characterized neuron subtype (see e). d, Two bona fide subclasses of Npas1+ neurons are identified in the mouse GPe. They differ in their molecular marker expression, axonal projections, and electrophysiological properties. Npas1+-Foxp2+ neurons represent 60% of the total Npas1+ neuron class; they project exclusively to the dStr, targeting SPNs. Npas1+-Npr3+ (also known as Npas1+-Nkx2.1+ and Npas1+-Lhx6+) neurons represent 40% of the total Npas1+ neuron class; they project to the cortex, thalamus, and midbrain. Npas1+-Foxp2+ neurons alone are sufficient to account for the motor-suppressing effect produced by optogenetic stimulation of pan-Npas1+ neurons in vivo. In contrast, the behavioral roles of Npas1+-Npr3+ awaits further clarification. Size of the target areas (circles) is an artistic rendering based on the volume of the target areas, and does not reflect the axonal density, synaptic strength, or contacts formed by Npas1+ neuron subclasses. e, Lhx6+ neurons are heterogeneous. They encompass three different subpopulations of neurons, including those in the PV+ and Npas1+ neuron classes. Both PV+-Lhx6+ neurons and Npas1+-Npr3+ neurons coexpress Sox6. Lhx6+-Sox6− neurons are a subset of PV−-Npas1− neurons.
Although cellular diversity in the GPe has been recognized since the 1970s (DeLong, 1971; Fox et al., 1974), how heterogeneity maps onto molecular and functional differences has been unclear. Our interest was never to understand cell type per se. Instead, by studying cellular composition and the nature of neuronal identity, we hope to uncover molecular markers and genetic tools that allow us to interrogate the organization principles and functions of GPe neuron subtypes. The cellular composition in the GPe is relatively simple compared with that of the cortex and hippocampus (Rudy et al., 2011; Jiang et al., 2015; Tasic et al., 2016, 2018; Huang and Paul, 2019; Adkins et al., 2020; Gouwens et al., 2020; Yao, 2020); the advent of single-cell transcriptomic and mouse genetics has created an opportunity to interrogate the GPe with previously unexplored tools and has enabled us to provide new insights into GPe neuron diversity and its behavioral relevance in this study (Fig. 9). A large set of transcription factors have been previously identified to govern the formation of the GPe. We took a leap of faith in generating Npas1-Cre mice and Npas1 antibodies de novo to interrogate the GPe. This decision was based on the compelling data from prior literature (Flandin et al., 2010; Nóbrega-Pereira et al., 2010). While PV-Cre and Npas1-Cre mice will continue to be instrumental to our research, we have now identified a set of transgenic lines that will allow the field to work together to solve more challenging questions and to reconcile discrepancies that arose from suboptimal tools.
Depending on the metric used, the definition of cell types differs. This is our first attempt to examine the cellular, circuit, and behavioral properties of Foxp2+ neurons, Npr3+ neurons, Kcng4+ neurons, and Npy2r+ neurons. This study is among the first to show that GPe neuron subclasses have specialized connections and functions (Fig. 9). However, we do not know the relationship between these neuron subtypes and the somatotopic organization of the circuit (Romanelli et al., 2005; Nambu, 2011; Bertino et al., 2020). In vivo optogenetics does not offer the resolution to answer this type of question. Our understanding of these neurons, and ultimately the GPe, will undoubtedly continue to be refined while we await driver lines that offer genetic access to the PV+-Kcng4– population.
Beyond go and no-go?
By tracking body kinematics and movement dynamics, we described how optogenetic manipulations of specific cell types gave rise to unique behavioral structures. With this information, we inferred the behavioral states of mice and functional interactions between individual neuron types. We found that, upon Npas1+ neuron stimulation, mice were not completely motionless. Instead, they spent more time executing fine movement, which on its own can be mediated by a complex circuit. As motionless and fine movement often co-occurred within a given time period, we interpret fine movement as a manifestation of action suppression. In turn, the basal ganglia are reset for selecting new actions: in this case, locomotion in an open-field arena.
As PV+ neurons and dSPNs converge onto the SNr (Grofova, 1975; Araki et al., 1985; Bolam et al., 1993; Mink, 1996; Smith et al., 1998; Connelly et al., 2010), it is conceivable that PV+ neurons and dSPNs are both involved in motor promotion. Anatomical studies show that SNr-projecting striatal neurons arborize within the GPe (Kawaguchi et al., 1990; Wu et al., 2000; Levesque and Parent, 2005; Fujiyama et al., 2011; Cui et al., 2021); however, the postsynaptic targets remain elusive. Our data suggest that they do not target PV+ neurons. Although the role of the basal ganglia in movement speed has long been recognized (Kornhuber, 1971; DeLong et al., 1984; Yttri and Dudman, 2016; Fobbs et al., 2020), here we argue that the basal ganglia control more than just go and no-go. As the basal ganglia are known to support a wide repertoire of innate complex behaviors (e.g., rearing and grooming) (Graybiel, 2008; Redgrave et al., 2010; Ahmari, 2016; Markowitz et al., 2018; Park et al., 2020), we should consider using movement dynamics broadly as metrics to measure basal ganglia function rather than relying solely on speed. Our experiments used different mouse strains to target selective neuron types for manipulations; therefore, it is rather difficult to untangle the bona fide cell-specific behavioral effects from nuanced differences in behavior across mouse strains. Future experiments that use intersectional approaches will yield more definitive answers.
We have recently shown that PV+ neurons and Npas1+ neurons are positioned differently within the basal ganglia. This inference is further supported by in vivo whole-cell analysis (Ketzef and Silberberg, 2020). We did not observe marked differences in the electrophysiological properties between neurons within each class. This implies that input-output transformation among them is similar. It is thus probable that neurons within each class receive inputs with similar physiological properties. In turn, both PV+ neurons and Npas1+ neurons exhibit complex projection patterns. It is now clear that this complexity is in part a result of a mixture of neuron subclasses (Mastro et al., 2014; Abdi et al., 2015; Dodson et al., 2015; Hernández et al., 2015; Fujiyama et al., 2016; Glajch et al., 2016; Hegeman et al., 2016; Abecassis et al., 2020). The formation of these “circuit modules” by neuron subclasses, in turn, dictates the network effects and thus behavioral outcome. We previously examined the synaptic interactions between GPe neurons and key structures within the basal ganglia: the dorsal striatum and the STN (Chan et al., 2004; Fan et al., 2012; Hernández et al., 2015; Cui et al., 2016, 2021; Glajch et al., 2016; Pamukcu et al., 2020). Given the diametric behavioral effects produced by these two populations of neurons, it was a logical next step to ask whether PV+ neuron and Npas1+ neurons are reciprocally inhibiting each other. In this study, by using a combination of transgenic lines, we have overcome some of the challenges associated with the interrogation of cell type-specific connectivity. Our data showed that PV+ neurons and Npas1+ neurons are connected to each other and thus support the idea that the diametric behavioral phenotype observed when one of these two classes of neurons was perturbed was a result of a push-pull function between PV+ neurons and Npas1+ neurons. We found that Foxp2+ neurons completely avoided PV+ neurons. More importantly, this observation suggests that input from Npas1+ neurons to PV+ neurons is provided by Npr3+ (also known as Npas1+-Nkx2.1+) neurons exclusively. This inference is consistent with the high density of local axons produced by Npr3+ neurons, as observed in Figure 1c. However, we have yet to uncover the properties of the connections made by Npr3+ neurons. In contrast, synaptic tagging experiments suggested that Kcng4+ neurons did not produce local inputs (Fig. 1c). It will be important to confirm this anatomical observation with functional studies. With the identification of new genetic tools, we should be in a position to study the synaptic connectivity and its alterations at greater depth.
Recent literature emphasized that PV+ neurons are at the intersection of motor-anxiety circuits (Sztainberg et al., 2011; Hunt et al., 2018; Giovanniello et al., 2020); we do not currently know the downstream targets that mediate the anxiety-related response. As the STN does not mediate this anxiety-related response, other descending pathways, such as the direct projections to the raphe (Pollak Dorocic et al., 2014) or their indirect projection to the habenula via the internal globus pallidus (Smith et al., 1998; Wallace et al., 2017), could be involved. While Npas1+ neurons, in particular Foxp2+ neurons, have extensive axonal arborization in the dorsal striatum (Dodson et al., 2015; Hernández et al., 2015; Glajch et al., 2016), t is important to emphasize that their downstream impact on the somatic excitability of striatal projection neurons is relatively weak (Glajch et al., 2016). The cellular mechanisms that mediate motor-suppression have yet to be identified. While Foxp2+ neurons have been proposed to send “stop” signals to the striatum (Mallet et al., 2016; Aristieta et al., 2020; Goenner et al., 2020), it is likely that multiple circuit elements are involved in producing complete behavioral arrest (Schmidt et al., 2013; Jahanshahi et al., 2015; Wessel and Aron, 2017). On the other hand, it is clear that the inhibitory projection to the STN (and substantial pars reticulata) mediates the motor-promoting effects, and it is consistent with the established relationship between STN activity and movement suppression (Hamani et al., 2004; Aron and Poldrack, 2006; Aron et al., 2007; Eagle et al., 2008; Schmidt et al., 2013; Schweizer et al., 2014; Fife et al., 2017; Wessel and Aron, 2017; Adam et al., 2020). In addition to the established literature demonstrating that the GPe sends inhibitory signals to the input and output nuclei of the basal ganglia (Jessell et al., 1978; Mink, 1996; Smith et al., 1998; Kita, 2007; Hegeman et al., 2016), recent studies highlight the existence of cortico-pallido-cortical loops (Naito and Kita, 1994; M. C. Chen et al., 2015; Saunders et al., 2015; Schwarz et al., 2015; Ahrlund-Richter et al., 2019; Karube et al., 2019; Abecassis et al., 2020; Adkins et al., 2020; Anastasiades et al., 2020; Clayton et al., 2020; Gehrlach et al., 2020; Lee et al., 2020; Muñoz-Castañeda, 2020; Garcia et al., 2021). However, the precise circuitry has yet to be defined. The direct feedback from Npr3+ (also known as Npas1+-Nkx2.1+) neurons to cortical regions (Fig. 1c) (see also Abecassis et al., 2020) argues that action selection is not simply a top-down command.
Footnotes
This work was supported by National Institutes of Health R01 NS069777, R01 MH112768, R01 NS097901, R01 MH109466, and R01 NS088528 to C.S.C., T32 AG020506 to A.P., T32 NS041234 and F32 NS098793 to H.S.X., R01 MH112768 to N.J.J., R35 NS097185 to C.J.W., and HHMI-PF Medical Research Fellowship and AΩA Student Research Fellowship to Z.A.A. We thank Daniela Garcia, Alyssa Bebenek, Moises Melesio, Kris Shah, Ahana Narayanan, Vaishnavi Tetali, Saivasudha Chalasani, and Daniel Hegeman for assistance on the project; Dr. Tiffany Schmidt for providing Kcng4-Cre mice; Dr. Byung Kook Lim for providing the synaptophysin-GFP virus; and Drs. Ann Kennedy and Sam Golden for input on the behavioral analyses.
The authors declare no competing financial interests.
References
- Abdi A, Mallet N, Mohamed FY, Sharott A, Dodson PD, Nakamura KC, Suri S, Avery SV, Larvin JT, Garas FN, Garas SN, Vinciati F, Morin S, Bezard E, Baufreton J, Magill PJ (2015) Prototypic and arkypallidal neurons in the dopamine-intact external globus pallidus. J Neurosci 35:6667–6688. 10.1523/JNEUROSCI.4662-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecassis ZA, Berceau BL, Win PH, Garcia D, Xenias HS, Cui Q, Pamukcu A, Cherian S, Hernández VM, Chon U, Lim BK, Kim Y, Justice NJ, Awatramani R, Hooks BM, Gerfen CR, Boca SM, Chan CS (2020) Npas1(+)-Nkx2.1(+) neurons are an integral part of the cortico-pallido-cortical loop. J Neurosci 40:743–768. 10.1523/JNEUROSCI.1199-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam EM, Johns T, Sur M (2020) Cortico-subthalamic projections send brief stop signals to halt visually-guided locomotion. bioRxiv 2020.2002.2005.936443. [Google Scholar]
- Adkins RS, Aldridge AI, Allen S, Ament SA, An X, Armand E, Ascoli GA, Bakken TE, Bandrowski A, Banerjee S, Barkas N, Bartlett A, Bateup HS, Behrens MM, Berens P, Berg J, Bernabucci M, Bernaerts Y, Bertagnolli D, Biancalani T, et al. (2020) A multimodal cell census and atlas of the mammalian primary motor cortex. bioRxiv. doi: 10.1101/2020.10.19.343129. [DOI] [Google Scholar]
- Ahmari SE (2016) Using mice to model obsessive compulsive disorder: from genes to circuits. Neuroscience 321:121–137. 10.1016/j.neuroscience.2015.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrlund-Richter S, Xuan Y, van Lunteren JA, Kim H, Ortiz C, Pollak Dorocic I, Meletis K, Carlén M (2019) A whole-brain atlas of monosynaptic input targeting four different cell types in the medial prefrontal cortex of the mouse. Nat Neurosci 22:657–668. 10.1038/s41593-019-0354-y [DOI] [PubMed] [Google Scholar]
- Albin RL, Young AB, Penney JB (1989) The functional anatomy of basal ganglia disorders. Trends Neurosci 12:366–375. 10.1016/0166-2236(89)90074-x [DOI] [PubMed] [Google Scholar]
- Anastasiades PG, Collins DP, Carter AG (2020) Mediodorsal and ventromedial thalamus engage distinct L1 circuits in the prefrontal cortex. Neuron 109:314–330.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson ME, Horak FB (1985) Influence of the globus pallidus on arm movements in monkeys: III. Timing of movement-related information. J Neurophysiol 54:433–448. 10.1152/jn.1985.54.2.433 [DOI] [PubMed] [Google Scholar]
- Araki M, McGeer PL, McGeer EG (1985) Striatonigral and pallidonigral pathways studied by a combination of retrograde horseradish peroxidase tracing and a pharmacohistochemical method for gamma-aminobutyric acid transaminase. Brain Res 331:17–24. 10.1016/0006-8993(85)90710-3 [DOI] [PubMed] [Google Scholar]
- Aristieta A, Barresi M, Azizpour LS, Barriere G, Courtand G, de la Crompe B, Guilhemsang L, Gauthier S, Fioramonti S, Baufreton J, Mallet NP (2020) A disynaptic circuit in the globus pallidus controls locomotion inhibition. Curr Biol 31:707–721.e7. [DOI] [PubMed] [Google Scholar]
- Aron AR, Poldrack RA (2006) Cortical and subcortical contributions to Stop signal response inhibition: role of the subthalamic nucleus. J Neurosci 26:2424–2433. 10.1523/JNEUROSCI.4682-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron AR, Durston S, Eagle DM, Logan GD, Stinear CM, Stuphorn V (2007) Converging evidence for a fronto-basal-ganglia network for inhibitory control of action and cognition. J Neurosci 27:11860–11864. 10.1523/JNEUROSCI.3644-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertino S, Basile GA, Bramanti A, Anastasi GP, Quartarone A, Milardi D, Cacciola A (2020) Spatially coherent and topographically organized pathways of the human globus pallidus. Hum Brain Mapp 41:4641–4661. 10.1002/hbm.25147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan MD, Booth PA, Eaton SA, Bolam JP (1998) Selective innervation of neostriatal interneurons by a subclass of neuron in the globus pallidus of the rat. J Neurosci 18:9438–9452. 10.1523/JNEUROSCI.18-22-09438.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolam JP, Smith Y, Ingham CA, von Krosigk M, Smith AD (1993) Convergence of synaptic terminals from the striatum and the globus pallidus onto single neurones in the substantia nigra and the entopeduncular nucleus. Prog Brain Res 99:73–88. 10.1016/s0079-6123(08)61339-4 [DOI] [PubMed] [Google Scholar]
- Boraud T, Bezard E, Guehl D, Bioulac B, Gross C (1998) Effects of L-DOPA on neuronal activity of the globus pallidus externalis (GPe) and globus pallidus internalis (GPi) in the MPTP-treated monkey. Brain Res 787:157–160. 10.1016/s0006-8993(97)01563-1 [DOI] [PubMed] [Google Scholar]
- Bugaysen J, Bar-Gad I, Korngreen A (2013) Continuous modulation of action potential firing by a unitary GABAergic connection in the globus pallidus in vitro. J Neurosci 33:12805–12809. 10.1523/JNEUROSCI.1970-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Shigemoto R, Mercer JN, Surmeier DJ (2004) HCN2 and HCN1 channels govern the regularity of autonomous pacemaking and synaptic resetting in globus pallidus neurons. J Neurosci 24:9921–9932. 10.1523/JNEUROSCI.2162-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Glajch KE, Gertler TS, Guzman JN, Mercer JN, Lewis AS, Goldberg AB, Tkatch T, Shigemoto R, Fleming SM, Chetkovich DM, Osten P, Kita H, Surmeier DJ (2011) HCN channelopathy in external globus pallidus neurons in models of Parkinson's disease. Nat Neurosci 14:85–92. 10.1038/nn.2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Peterson JD, Gertler TS, Glajch KE, Quintana RE, Cui Q, Sebel LE, Plotkin JL, Shen W, Heiman M, Heintz N, Greengard P, Surmeier DJ (2012) Strain-specific regulation of striatal phenotype in Drd2-eGFP BAC transgenic mice. J Neurosci 32:9124–9132. 10.1523/JNEUROSCI.0229-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang RB, Strochlic DE, Williams EK, Umans BD, Liberles SD (2015) Vagal sensory neuron subtypes that differentially control breathing. Cell 161:622–633. 10.1016/j.cell.2015.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MC, Ferrari L, Sacchet MD, Foland-Ross LC, Qiu MH, Gotlib IH, Fuller PM, Arrigoni E, Lu J (2015) Identification of a direct GABAergic pallidocortical pathway in rodents. Eur J Neurosci 41:748–759. 10.1111/ejn.12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Friedman BA, Ha C, Durinck S, Liu J, Rubenstein JL, Seshagiri S, Modrusan Z (2017) Single-cell RNA sequencing identifies distinct mouse medial ganglionic eminence cell types. Sci Rep 7:45656. 10.1038/srep45656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton KK, Williamson RS, Hancock KE, Tasaka GI, Mizrahi A, Hackett TA, Polley DB (2020) Auditory corticothalamic neurons are recruited by motor preparatory inputs. Curr Biol 31:310–321.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly WM, Schulz JM, Lees G, Reynolds JN (2010) Differential short-term plasticity at convergent inhibitory synapses to the substantia nigra pars reticulata. J Neurosci 30:14854–14861. 10.1523/JNEUROSCI.3895-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Pitt JE, Pamukcu A, Poulin JF, Mabrouk OS, Fiske MP, Fan IB, Augustine EC, Young KA, Kennedy RT, Awatramani R, Chan CS (2016) Blunted mGluR activation disinhibits striatopallidal transmission in Parkinsonian mice. Cell Rep 17:2431–2444. 10.1016/j.celrep.2016.10.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Du X, Chang IY, Pamukcu A, Lilascharoen V, Berceau BL, García D, Hong D, Chon U, Narayanan A, Kim Y, Lim BK, Chan CS (2021) Striatal direct pathway targets Npas1+ pallidal neurons. J Neurosci 41:3966–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen RS, Takeno MM, Huang L, Gu H, Larsen R, Mills M, Bosma-Moody A, Siverts LA, Walker M, Graybuck LT, Yao Z, Fong O, Nguyen TN, Garren E, Lenz GH, et al. (2018) A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell 174:465–480.e422. 10.1016/j.cell.2018.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehorter N, Ciceri G, Bartolini G, Lim L, del Pino I, Marín O (2015) Tuning of fast-spiking interneuron properties by an activity-dependent transcriptional switch. Science 349:1216–1220. 10.1126/science.aab3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong MR (1971) Activity of pallidal neurons during movement. J Neurophysiol 34:414–427. 10.1152/jn.1971.34.3.414 [DOI] [PubMed] [Google Scholar]
- DeLong MR (1990) Primate models of movement disorders of basal ganglia origin. Trends Neurosci 13:281–285. 10.1016/0166-2236(90)90110-v [DOI] [PubMed] [Google Scholar]
- DeLong MR, Wichmann T (2007) Circuits and circuit disorders of the basal ganglia. Arch Neurol 64:20–24. 10.1001/archneur.64.1.20 [DOI] [PubMed] [Google Scholar]
- DeLong MR, Alexander GE, Georgopoulos AP, Crutcher MD, Mitchell SJ, Richardson RT (1984) Role of basal ganglia in limb movements. Hum Neurobiol 2:235–244. [PubMed] [Google Scholar]
- Dodson PD, Larvin JT, Duffell JM, Garas FN, Doig NM, Kessaris N, Duguid IC, Bogacz R, Butt SJ, Magill PJ (2015) Distinct developmental origins manifest in the specialized encoding of movement by adult neurons of the external globus pallidus. Neuron 86:501–513. 10.1016/j.neuron.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, De la Huerta I, Sanes JR (2014) Type II cadherins guide assembly of a direction-selective retinal circuit. Cell 158:793–807. 10.1016/j.cell.2014.06.047 [DOI] [PubMed] [Google Scholar]
- Dudman JT, Krakauer JW (2016) The basal ganglia: from motor commands to the control of vigor. Curr Opin Neurobiol 37:158–166. 10.1016/j.conb.2016.02.005 [DOI] [PubMed] [Google Scholar]
- Eagle DM, Baunez C, Hutcheson DM, Lehmann O, Shah AP, Robbins TW (2008) Stop-signal reaction-time task performance: role of prefrontal cortex and subthalamic nucleus. Cereb Cortex 18:178–188. 10.1093/cercor/bhm044 [DOI] [PubMed] [Google Scholar]
- Evans RC, Twedell EL, Zhu M, Ascencio J, Zhang R, Khaliq ZM (2020) Functional dissection of basal ganglia inhibitory inputs onto substantia nigra dopaminergic neurons. Cell Rep 32:108156. 10.1016/j.celrep.2020.108156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan KY, Baufreton J, Surmeier DJ, Chan CS, Bevan MD (2012) Proliferation of external globus pallidus-subthalamic nucleus synapses following degeneration of midbrain dopamine neurons. J Neurosci 32:13718–13728. 10.1523/JNEUROSCI.5750-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fife KH, Gutierrez-Reed NA, Zell V, Bailly J, Lewis CM, Aron AR, Hnasko TS (2017) Causal role for the subthalamic nucleus in interrupting behavior. Elife 6:e27689. 10.7554/eLife.27689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion M, Tremblay L, Bedard PJ (1991) Effects of dopamine agonists on the spontaneous activity of globus pallidus neurons in monkeys with MPTP-induced parkinsonism. Brain Res 547:152–161. [PubMed] [Google Scholar]
- Fishell G, Kepecs A (2020) Interneuron types as attractors and controllers. Annu Rev Neurosci 43:1–30. 10.1146/annurev-neuro-070918-050421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Kimura S, Rubenstein JL (2010) The progenitor zone of the ventral medial ganglionic eminence requires Nkx2-1 to generate most of the globus pallidus but few neocortical interneurons. J Neurosci 30:2812–2823. 10.1523/JNEUROSCI.4228-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobbs WC, Bariselli S, Licholai JA, Miyazaki NL, Matikainen-Ankney BA, Creed MC, Kravitz AV (2020) Continuous representations of speed by striatal medium spiny neurons. J Neurosci 40:1679–1688. 10.1523/JNEUROSCI.1407-19.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CA, Andrade AN, Lu Qui IJ, Rafols JA (1974) The primate globus pallidus: a Golgi and electron microscopic study. J Hirnforsch 15:75–93. [PubMed] [Google Scholar]
- Fujiyama F, Sohn J, Nakano T, Furuta T, Nakamura KC, Matsuda W, Kaneko T (2011) Exclusive and common targets of neostriatofugal projections of rat striosome neurons: a single neuron-tracing study using a viral vector. Eur J Neurosci 33:668–677. 10.1111/j.1460-9568.2010.07564.x [DOI] [PubMed] [Google Scholar]
- Fujiyama F, Nakano T, Matsuda W, Furuta T, Udagawa J, Kaneko T (2016) A single-neuron tracing study of arkypallidal and prototypic neurons in healthy rats. Brain Struct Funct 221:4733–4740. 10.1007/s00429-015-1152-2 [DOI] [PubMed] [Google Scholar]
- Garas FN, Shah RS, Kormann E, Doig NM, Vinciati F, Nakamura KC, Dorst MC, Smith Y, Magill PJ, Sharott A (2016) Secretagogin expression delineates functionally-specialized populations of striatal parvalbumin-containing interneurons. Elife 5:e16088. 10.7554/eLife.16088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AF, Crummy EA, Webb IG, Nooney MN, Ferguson SM (2021) Distinct populations of cortical pyramidal neurons mediate drug reward and aversion. Nat Commun 12:182. 10.1038/s41467-020-20526-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrlach DA, Weiand C, Gaitanos TN, Cho E, Klein AS, Hennrich AA, Conzelmann KK, Gogolla N (2020) A whole-brain connectivity map of mouse insular cortex. Elife 9:e55585. 10.7554/eLife.55585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman D, Griveau A, Dehorter N, Teissier A, Varela C, Pla R, Pierani A, Marin O (2011) A wide diversity of cortical GABAergic interneurons derives from the embryonic preoptic area. J Neurosci 31:16570–16580. 10.1523/JNEUROSCI.4068-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanniello J, Yu K, Furlan A, Nachtrab GT, Sharma R, Chen X, Li B (2020) A central amygdala-globus pallidus circuit conveys unconditioned stimulus-related information and controls fear learning. J Neurosci 40:9043–9054. 10.1523/JNEUROSCI.2090-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glajch KE, Kelver DA, Hegeman DJ, Cui Q, Xenias HS, Augustine EC, Hernández VM, Verma N, Huang TY, Luo M, Justice NJ, Chan CS (2016) Npas1+ pallidal neurons target striatal projection neurons. J Neurosci 36:5472–5488. 10.1523/JNEUROSCI.1720-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenner L, Maith O, Koulouri I, Baladron J, Hamker FH (2020) A spiking model of basal ganglia dynamics in stopping behavior supported by arkypallidal neurons. Eur J Neurosci Advance online publication. Retrieved Dec 14, 2020. doi: 10.1111/ejn.15082. [DOI] [PubMed] [Google Scholar]
- Gouwens NW, Sorensen SA, Baftizadeh F, Budzillo A, Lee BR, Jarsky T, Alfiler L, Baker K, Barkan E, Berry K, Bertagnolli D, Bickley K, Bomben J, Braun T, Brouner K, Casper T, Crichton K, Daigle TL, Dalley R, de Frates RA, et al. (2020) Integrated morphoelectric and transcriptomic classification of cortical GABAergic cells. Cell 183:935–953.e919. 10.1016/j.cell.2020.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybiel AM (2008) Habits, rituals, and the evaluative brain. Annu Rev Neurosci 31:359–387. 10.1146/annurev.neuro.29.051605.112851 [DOI] [PubMed] [Google Scholar]
- Graybiel AM, Aosaki T, Flaherty AW, Kimura M (1994) The basal ganglia and adaptive motor control. Science 265:1826–1831. 10.1126/science.8091209 [DOI] [PubMed] [Google Scholar]
- Grillner S, Robertson B (2016) The basal ganglia over 500 million years. Curr Biol 26:R1088–R1100. 10.1016/j.cub.2016.06.041 [DOI] [PubMed] [Google Scholar]
- Grofova I (1975) The identification of striatal and pallidal neurons projecting to substantia nigra: an experimental study by means of retrograde axonal transport of horseradish peroxidase. Brain Res 91:286–291. [DOI] [PubMed] [Google Scholar]
- Gu BM, Schmidt R, Berke JD (2020) Globus pallidus dynamics reveal covert strategies for behavioral inhibition. Elife 9:e57215. 10.7554/eLife.57215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A (2011) Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci 14:1481–1488. 10.1038/nn.2928 [DOI] [PubMed] [Google Scholar]
- Hamani C, Saint-Cyr JA, Fraser J, Kaplitt M, Lozano AM (2004) The subthalamic nucleus in the context of movement disorders. Brain 127:4–20. 10.1093/brain/awh029 [DOI] [PubMed] [Google Scholar]
- Hegeman DJ, Hong ES, Hernández VM, Chan CS (2016) The external globus pallidus: progress and perspectives. Eur J Neurosci 43:1239–1265. 10.1111/ejn.13196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández VM, Hegeman DJ, Cui Q, Kelver DA, Fiske MP, Glajch KE, Pitt JE, Huang TY, Justice NJ, Chan CS (2015) Parvalbumin+ neurons and Npas1+ neurons are distinct neuron classes in the mouse external globus pallidus. J Neurosci 35:11830–11847. 10.1523/JNEUROSCI.4672-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs MH, Wilson CJ (2016) Unitary synaptic connections among substantia nigra pars reticulata neurons. J Neurophysiol 115:2814–2829. 10.1152/jn.00094.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, Yao Z, Eggermont J, Höllt T, Levi BP, Shehata SI, Aevermann B, Beller A, Bertagnolli D, Brouner K, Casper T, et al. (2019) Conserved cell types with divergent features in human versus mouse cortex. Nature 573:61–68. 10.1038/s41586-019-1506-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JS, Vogt D, Sandberg M, Rubenstein JL (2017) Cortical interneuron development: a tale of time and space. Development 144:3867–3878. 10.1242/dev.132852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, Paul A (2019) The diversity of GABAergic neurons and neural communication elements. Nat Rev Neurosci 20:563–572. 10.1038/s41583-019-0195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AJ Jr, Dasgupta R, Rajamanickam S, Jiang Z, Beierlein M, Chan CS, Justice NJ (2018) Paraventricular hypothalamic and amygdalar CRF neurons synapse in the external globus pallidus. Brain Struct Funct 223:2685–2698. 10.1007/s00429-018-1652-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison WD, Lozano AM, Davis KD, Saint-Cyr JA, Lang AE, Dostrovsky JO (1994) Differential neuronal activity in segments of globus pallidus in Parkinson's disease patients. Neuroreport 5:1533–1537. 10.1097/00001756-199407000-00031 [DOI] [PubMed] [Google Scholar]
- Jaeger D, Kita H (2011) Functional connectivity and integrative properties of globus pallidus neurons. Neuroscience 198:44–53. 10.1016/j.neuroscience.2011.07.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshahi M, Obeso I, Rothwell JC, Obeso JA (2015) A fronto-striato-subthalamic-pallidal network for goal-directed and habitual inhibition. Nat Rev Neurosci 16:719–732. 10.1038/nrn4038 [DOI] [PubMed] [Google Scholar]
- Jessell TM, Emson PC, Paxinos G, Cuello AC (1978) Topographic projections of substance P and GABA pathways in the striato- and pallido-nigral system: a biochemical and immunohistochemical study. Brain Res 152:487–498. 10.1016/0006-8993(78)91104-6 [DOI] [PubMed] [Google Scholar]
- Jiang X, Shen S, Cadwell CR, Berens P, Sinz F, Ecker AS, Patel S, Tolias AS (2015) Principles of connectivity among morphologically defined cell types in adult neocortex. Science 350:aac9462. 10.1126/science.aac9462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Tecuapetla F, Costa RM (2014) Basal ganglia subcircuits distinctively encode the parsing and concatenation of action sequences. Nat Neurosci 17:423–430. 10.1038/nn.3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karube F, Takahashi S, Kobayashi K, Fujiyama F (2019) Motor cortex can directly drive the globus pallidus neurons in a projection neuron type-dependent manner in the rat. Elife 8:e49511. 10.7554/eLife.49511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Wilson CJ, Emson PC (1990) Projection subtypes of rat neostriatal matrix cells revealed by intracellular injection of biocytin. J Neurosci 10:3421–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketzef M, Silberberg G (2020) Differential synaptic input to external globus pallidus neuronal subpopulations in vivo. Neuron 109:516–529.e4. [DOI] [PubMed] [Google Scholar]
- Kita H (1994) Parvalbumin-immunopositive neurons in rat globus pallidus: a light and electron microscopic study. Brain Res 657:31–41. 10.1016/0006-8993(94)90950-4 [DOI] [PubMed] [Google Scholar]
- Kita H (2007) Globus pallidus external segment. Prog Brain Res 160:111–133. 10.1016/S0079-6123(06)60007-1 [DOI] [PubMed] [Google Scholar]
- Kita H, Kitai ST (1994) The morphology of globus pallidus projection neurons in the rat: an intracellular staining study. Brain Res 636:308–319. 10.1016/0006-8993(94)91030-8 [DOI] [PubMed] [Google Scholar]
- Klaus A, Alves da Silva J, Costa RM (2019) What, if, and when to move: basal ganglia circuits and self-paced action initiation. Annu Rev Neurosci 42:459–483. 10.1146/annurev-neuro-072116-031033 [DOI] [PubMed] [Google Scholar]
- Kornhuber HH (1971) Motor functions of cerebellum and basal ganglia: the cerebellocortical saccadic (ballistic) clock, the cerebellonuclear hold regulator, and the basal ganglia ramp (voluntary speed smooth movement) generator. Kybernetik 8:157–162. 10.1007/BF00290561 [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Altman N (2014) Visualizing samples with box plots. Nat Methods 11:119–120. 10.1038/nmeth.2813 [DOI] [PubMed] [Google Scholar]
- Lee J, Wang W, Sabatini BL (2020) Anatomically segregated basal ganglia pathways allow parallel behavioral modulation. Nat Neurosci 23:1388–1398. 10.1038/s41593-020-00712-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Levesque M, Parent A (2005) The striatofugal fiber system in primates: a reevaluation of its organization based on single-axon tracing studies. Proc Natl Acad Sci USA 102:11888–11893. 10.1073/pnas.0502710102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leys C, Ley C, Klein O, Bernard P, Licata L (2013) Detecting outliers: do not use standard deviation around the mean, use absolute deviation around the median. J Exp Soc Psychol 49:764–766. 10.1016/j.jesp.2013.03.013 [DOI] [Google Scholar]
- Lim L, Mi D, Llorca A, Marin O (2018) Development and functional diversification of cortical interneurons. Neuron 100:294–313. 10.1016/j.neuron.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill PJ, Bolam JP, Bevan MD (2001) Dopamine regulates the impact of the cerebral cortex on the subthalamic nucleus-globus pallidus network. Neuroscience 106:313–330. 10.1016/s0306-4522(01)00281-0 [DOI] [PubMed] [Google Scholar]
- Mahadevan V, Mitra A, Zhang Y, Peltekian A, Chittajallu R, Esnault C, Maric D, Rhodes C, Pelkey KA, Dale R, Petros TJ, McBain CJ (2020) NMDAR-mediated transcriptional control of gene expression in the specification of interneuron subtype identity. bioRxiv 2020.2006.2010.144295. [Google Scholar]
- Mallet N, Pogosyan A, Marton LF, Bolam JP, Brown P, Magill PJ (2008) Parkinsonian beta oscillations in the external globus pallidus and their relationship with subthalamic nucleus activity. J Neurosci 28:14245–14258. 10.1523/JNEUROSCI.4199-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet N, Schmidt R, Leventhal D, Chen F, Am N, Boraud T, Berke JD (2016) Arkypallidal cells send a stop signal to striatum. Neuron 89:308–316. 10.1016/j.neuron.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz JE, Gillis WF, Beron CC, Neufeld SQ, Robertson K, Bhagat ND, Peterson RE, Peterson E, Hyun M, Linderman SW, Sabatini BL, Datta SR (2018) The striatum organizes 3D behavior via moment-to-moment action selection. Cell 174:44–58.e17. 10.1016/j.cell.2018.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastro KJ, Bouchard RS, Holt HA, Gittis AH (2014) Transgenic mouse lines subdivide external segment of the globus pallidus (GPe) neurons and reveal distinct GPe output pathways. J Neurosci 34:2087–2099. 10.1523/JNEUROSCI.4646-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastro KJ, Zitelli KT, Willard AM, Leblanc KH, Kravitz AV, Gittis AH (2017) Cell-specific pallidal intervention induces long-lasting motor recovery in dopamine-depleted mice. Nat Neurosci 20:815–823. 10.1038/nn.4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis A, Yüksekgönül M, Rogers B, Bethge M, Mathis MW (2019) Pretraining boosts out-of-domain robustness for pose estimation. arXiv 1909.11229. [Google Scholar]
- Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, Mathis MW, Bethge M (2018) DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat Neurosci 21:1281–1289. 10.1038/s41593-018-0209-y [DOI] [PubMed] [Google Scholar]
- Metsalu T, Vilo J (2015) ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res 43:W566–W570. 10.1093/nar/gkv468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink JW (1996) The basal ganglia: focused selection and inhibition of competing motor programs. Prog Neurobiol 50:381–425. 10.1016/s0301-0082(96)00042-1 [DOI] [PubMed] [Google Scholar]
- Muñoz-Castañeda R (2020) Cellular anatomy of the mouse primary motor cortex. bioRxiv 2020.2010.2002.323154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito A, Kita H (1994) The cortico-pallidal projection in the rat: an anterograde tracing study with biotinylated dextran amine. Brain Res 653:251–257. 10.1016/0006-8993(94)90397-2 [DOI] [PubMed] [Google Scholar]
- Nambu A (2011) Somatotopic organization of the primate basal ganglia. Front Neuroanat 5:26. 10.3389/fnana.2011.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu A, Llinás R (1997) Morphology of globus pallidus neurons: its correlation with electrophysiology in guinea pig brain slices. J Comp Neurol 377:85–94. [DOI] [PubMed] [Google Scholar]
- Nath T, Mathis A, Chen AC, Patel A, Bethge M, Mathis MW (2019) Using DeepLabCut for 3D markerless pose estimation across species and behaviors. Nat Protoc 14:2152–2176. 10.1038/s41596-019-0176-0 [DOI] [PubMed] [Google Scholar]
- Nelson AB, Kreitzer AC (2014) Reassessing models of basal ganglia function and dysfunction. Annu Rev Neurosci 37:117–135. 10.1146/annurev-neuro-071013-013916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson SR, Goodwin NL, Choong JJ, Hwang S, Wright HR, Norville ZC, Tong X, Lin D, Bentzley BS, Eshel N, McLaughlin RJ, Golden SA (2020) Simple Behavioral Analysis (SimBA): an open source toolkit for computer classification of complex social behaviors in experimental animals. bioRxiv 2020.2004.2019.049452. [Google Scholar]
- Nini A, Feingold A, Slovin H, Bergman H (1995) Neurons in the globus pallidus do not show correlated activity in the normal monkey, but phase-locked oscillations appear in the MPTP model of parkinsonism. J Neurophysiol 74:1800–1805. 10.1152/jn.1995.74.4.1800 [DOI] [PubMed] [Google Scholar]
- Nóbrega-Pereira S, Gelman D, Bartolini G, Pla R, Pierani A, Marín O (2010) Origin and molecular specification of globus pallidus neurons. J Neurosci 30:2824–2834. 10.1523/JNEUROSCI.4023-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzzo RL (2016) The box plots alternative for visualizing quantitative data. PM R 8:268–272. 10.1016/j.pmrj.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Pamukcu A, Cui Q, Xenias HS, Berceau BL, Augustine EC, Fan I, Chalasani S, Hantman AW, Lerner TN, Boca SM, Chan CS (2020) Parvalbumin(+) and Npas1(+) pallidal neurons have distinct circuit topology and function. J Neurosci 40:7855–7876. 10.1523/JNEUROSCI.0361-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Coddington LT, Dudman JT (2020) Basal ganglia circuits for action specification. Annu Rev Neurosci 43:485–507. 10.1146/annurev-neuro-070918-050452 [DOI] [PubMed] [Google Scholar]
- Pollak Dorocic I, Furth D, Xuan Y, Johansson Y, Pozzi L, Silberberg G, Carlen M, Meletis K (2014) A whole-brain atlas of inputs to serotonergic neurons of the dorsal and median raphe nuclei. Neuron 83:663–678. 10.1016/j.neuron.2014.07.002 [DOI] [PubMed] [Google Scholar]
- Poulin JF, Luppi MP, Hofer C, Caronia G, Hsu PK, Chan CS, Awatramani R (2020) PRISM: a progenitor-restricted intersectional fate mapping approach redefines forebrain lineages. Dev Cell 53:740–753.e743. 10.1016/j.devcel.2020.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz A, Vaadia E, Bergman H (2000) Firing patterns and correlations of spontaneous discharge of pallidal neurons in the normal and the tremulous 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine vervet model of parkinsonism. J Neurosci 20:8559–8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redgrave P, Rodriguez M, Smith Y, Rodriguez-Oroz MC, Lehericy S, Bergman H, Agid Y, DeLong MR, Obeso JA (2010) Goal-directed and habitual control in the basal ganglia: implications for Parkinson's disease. Nat Rev Neurosci 11:760–772. 10.1038/nrn2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanelli P, Esposito V, Schaal DW, Heit G (2005) Somatotopy in the basal ganglia: experimental and clinical evidence for segregated sensorimotor channels. Brain Res Brain Res Rev 48:112–128. 10.1016/j.brainresrev.2004.09.008 [DOI] [PubMed] [Google Scholar]
- Rothblat DS, Schneider JS (1995) Alterations in pallidal neuronal responses to peripheral sensory and striatal stimulation in symptomatic and recovered parkinsonian cats. Brain Res 705:1–14. 10.1016/0006-8993(95)00892-6 [DOI] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J (2011) Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol 71:45–61. 10.1002/dneu.20853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadek AR, Magill PJ, Bolam JP (2007) A single-cell analysis of intrinsic connectivity in the rat globus pallidus. J Neurosci 27:6352–6362. 10.1523/JNEUROSCI.0953-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC (2018) MobileNetV2: inverted residuals and linear bottlenecks. arXiv 1801.04381. [Google Scholar]
- Sato F, Lavallée P, Lévesque M, Parent A (2000) Single-axon tracing study of neurons of the external segment of the globus pallidus in primate. J Comp Neurol 417:17–31. [DOI] [PubMed] [Google Scholar]
- Saunders A, Oldenburg IA, Berezovskii VK, Johnson CA, Kingery ND, Elliott HL, Xie T, Gerfen CR, Sabatini BL (2015) A direct GABAergic output from the basal ganglia to frontal cortex. Nature 521:85–89. 10.1038/nature14179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, Goeva A, Nemesh J, Kamitaki N, Brumbaugh S, Kulp D, McCarroll SA (2018) Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174:1015–1030.e1016. 10.1016/j.cell.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardoni G, Petterlini M, Laudanna C (2009) Analyzing biological network parameters with CentiScaPe. Bioinformatics 25:2857–2859. 10.1093/bioinformatics/btp517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Leventhal DK, Mallet N, Chen F, Berke JD (2013) Canceling actions involves a race between basal ganglia pathways. Nat Neurosci 16:1118–1124. 10.1038/nn.3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz LA, Miyamichi K, Gao XJ, Beier KT, Weissbourd B, DeLoach KE, Ren J, Ibanes S, Malenka RC, Kremer EJ, Luo L (2015) Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature 524:88–92. 10.1038/nature14600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer N, Pupe S, Arvidsson E, Nordenankar K, Smith-Anttila CJ, Mahmoudi S, Andrén A, Dumas S, Rajagopalan A, Lévesque D, Leão RN, Wallén-Mackenzie Å (2014) Limiting glutamate transmission in a Vglut2-expressing subpopulation of the subthalamic nucleus is sufficient to cause hyperlocomotion. Proc Natl Acad Sci USA 111:7837–7842. 10.1073/pnas.1323499111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LH, Luo F, Woodward DJ, Chang JY (2004) Neural responses in multiple basal ganglia regions during spontaneous and treadmill locomotion tasks in rats. Exp Brain Res 157:303–314. 10.1007/s00221-004-1844-y [DOI] [PubMed] [Google Scholar]
- Silberberg SN, Taher L, Lindtner S, Sandberg M, Nord AS, Vogt D, McKinsey GL, Hoch R, Pattabiraman K, Zhang D, Ferran JL, Rajkovic A, Golonzhka O, Kim C, Zeng H, Puelles L, Visel A, Rubenstein JL (2016) Subpallial enhancer transgenic lines: a data and tool resource to study transcriptional regulation of GABAergic cell fate. Neuron 92:59–74. 10.1016/j.neuron.2016.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Y, Bevan MD, Shink E, Bolam JP (1998) Microcircuitry of the direct and indirect pathways of the basal ganglia. Neuroscience 86:353–387. 10.1016/s0306-4522(98)00004-9 [DOI] [PubMed] [Google Scholar]
- Sousa VH, Fishell G (2010) Sonic hedgehog functions through dynamic changes in temporal competence in the developing forebrain. Curr Opin Genet Dev 20:391–399. 10.1016/j.gde.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit M, Gehlenborg N (2014) Bar charts and box plots. Nat Methods 11:117. 10.1038/nmeth.2807 [DOI] [PubMed] [Google Scholar]
- Sztainberg Y, Kuperman Y, Justice N, Chen A (2011) An anxiolytic role for CRF receptor type 1 in the globus pallidus. J Neurosci 31:17416–17424. 10.1523/JNEUROSCI.3087-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, Bertagnolli D, Goldy J, Shapovalova N, Parry S, Lee C, Smith K, Bernard A, Madisen L, Sunkin SM, Hawrylycz M, et al. (2016) Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19:335–346. 10.1038/nn.4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, Penn O, Bakken T, Menon V, Miller J, Fong O, Hirokawa KE, Lathia K, Rimorin C, Tieu M, Larsen R, et al. (2018) Shared and distinct transcriptomic cell types across neocortical areas. Nature 563:72–78. 10.1038/s41586-018-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RS, Anderson ME (2005) Context-dependent modulation of movement-related discharge in the primate globus pallidus. J Neurosci 25:2965–2976. 10.1523/JNEUROSCI.4036-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace ML, Saunders A, Huang KW, Philson AC, Goldman M, Macosko EZ, McCarroll SA, Sabatini BL (2017) Genetically distinct parallel pathways in the entopeduncular nucleus for limbic and sensorimotor output of the basal ganglia. Neuron 94:138–152.e135. 10.1016/j.neuron.2017.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel JR, Aron AR (2017) On the globality of motor suppression: unexpected events and their influence on behavior and cognition. Neuron 93:259–280. 10.1016/j.neuron.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Richard S, Parent A (2000) The organization of the striatal output system: a single-cell juxtacellular labeling study in the rat. Neurosci Res 38:49–62. 10.1016/s0168-0102(00)00140-1 [DOI] [PubMed] [Google Scholar]
- Yao Z (2020) A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. bioRxiv 2020.2003.2030.015214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yttri EA, Dudman JT (2016) Opponent and bidirectional control of movement velocity in the basal ganglia. Nature 533:402–406. 10.1038/nature17639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan XS, Wang L, Dong H, Qu WM, Yang SR, Cherasse Y, Lazarus M, Schiffmann SN, d'Exaerde AK, Li RX, Huang ZL (2017) Striatal adenosine A2A receptor neurons control active-period sleep via parvalbumin neurons in external globus pallidus. Elife 6:e29055. 10.7554/eLife.29055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimnik AJ, Nora GJ, Desmurget M, Turner RS (2015) Movement-related discharge in the macaque globus pallidus during high-frequency stimulation of the subthalamic nucleus. J Neurosci 35:3978–3989. 10.1523/JNEUROSCI.4899-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]