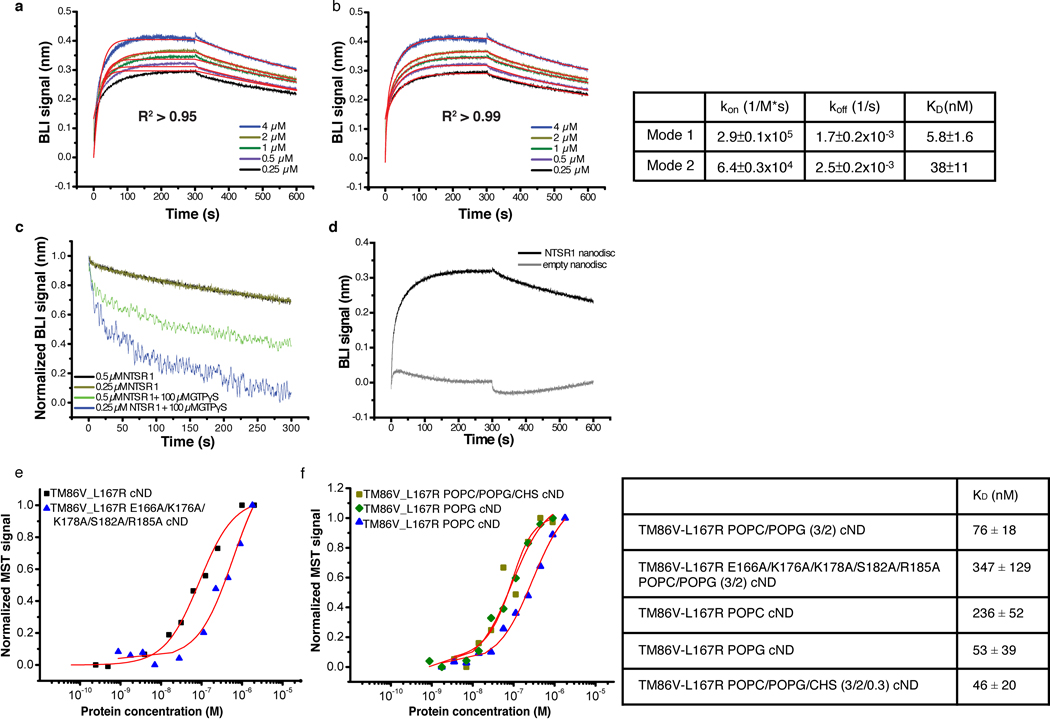
Extended Data Fig. 4 |. Characterization of the binding kinetics between NTS-NTSR1 and Gi in cNDs.
a-b, Fitting of Bio-Layer Interferometry (BLI) traces of Gi binding to NTS-NTSR1-cND using (a) one binding mode and (b) two binding mode shows better fitting using two binding mode. Right, a table showing kon, koff and KD from the two binding mode fitting. c, Dissociation between Gi and NTS-NTSR1-cND in the absence (black and brown) and presence (green and blue) of GTPγS, showing faster dissociation of the complex in the presence of GTPγS, suggesting that the NTSR1-Gαi1β1γ1 complex in cNDs is capable of GDP/GTP exchange. d, Association and dissociation kinetics of Gi binding to NTS-NTSR1-cND (dark) and empty cND (gray), showing much slower association and faster dissociation of Gi binding to empty cND compared to NTS-NTSR1-cND, suggesting that interaction between Gi and NTS-NTSR1-cND is driven by Gi binding to NTSR1 rather than to the nanodisc. e, Microscale thermophoresis (MST) data for the binding between NTSR1 and Gi (square mark), as well as the binding between mutant TM86V-L167R E166A/K176A/K178A/S182A/R185A and Gi (triangle mark) in POPC/POPG (3/2) cND. f, MST data for the binding between NTSR1 and Gi in POPC cND (triangle mark), POPG cND (diamond mark) and POPC/POPG/CHS cND (square mark). Right, a table showing KD from e-f.