Figure 4.
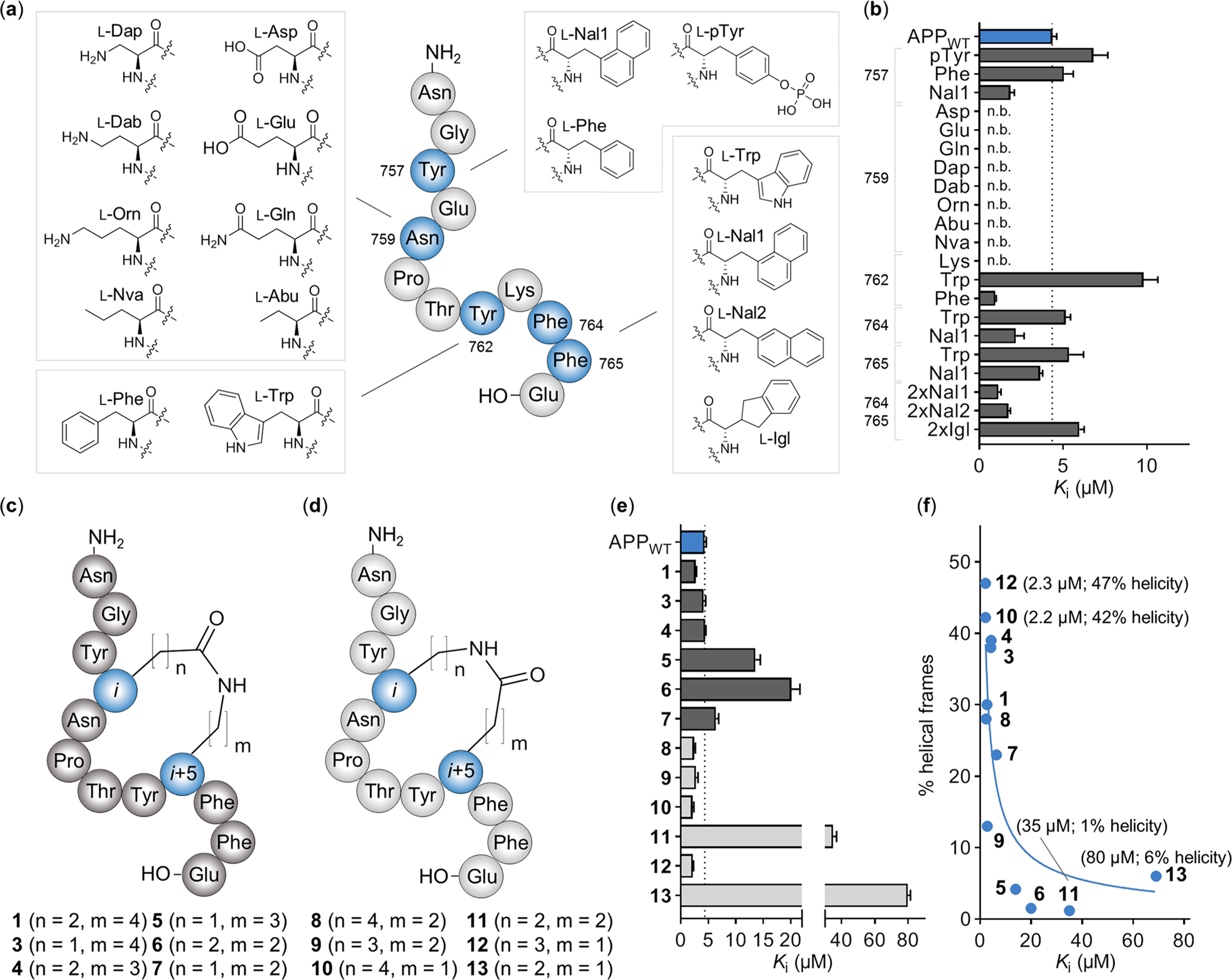
Incorporation of ncAAs and evaluation of side chain-to-side chain macrocyclization in the APPWT peptide. (a) Overview of synthesized APPWT peptide variants, including the structure of the introduced amino acids for each position (blue). (b) Ki values of each APPWT peptide variant measured by FP. n.b. indicates nonbinding peptide (i.e., Ki ≥ 500 μM). Data are expressed as the mean + SEM (n = 3). (c) Structure of side chain-to-side chain cyclized APPWT peptide analogues with native residue order (Glu in the i position, dark gray) and (d) inverse residue order (Lys in the i position, light gray). (e) Ki values of cyclic APPWT peptide variants measured by FP. Data are expressed as the mean + SEM (n = 3). (f) Helical propensity of residues Y762 to E766 from the molecular dynamics simulation of cyclic APPWT peptide variants 1 and 3–13 (% helical frames) plotted against the binding affinity (the mean Ki value) including the trend line (blue).
