Figure 7.
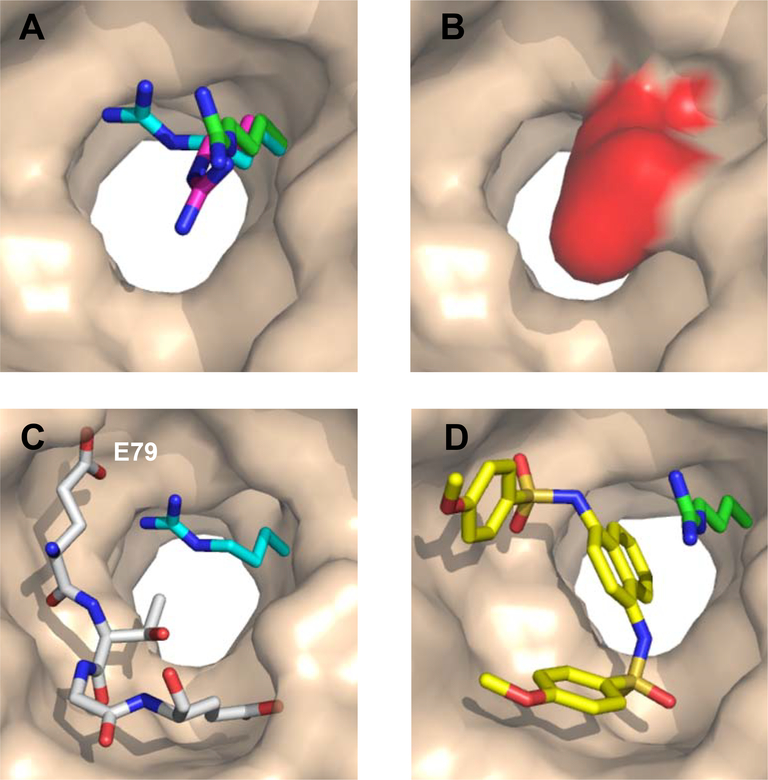
Three distinct conformations of Arg415 representative of those seen across different structures of KEAP1. (A) The positions seen for R415 in unbound KEAP1 (magenta, 4IFJ), in KEAP1 in complex with an Nrf2-derived 16-mer peptide encompassing the DxETGE motif (cyan, 4IFL; ligand not shown), and in KEAP1 in complex with a small-molecule ligand (green, 4IQK; ligand not shown). In wheat is the surface of unbound KEAP1 (4IFJ), omitting R415. (B) In unbound KEAP1, R415 (red) largely blocks access to the central channel at the base of the binding site. (C) In the peptide-bound KEAP1, the side-chain of R415 (cyan) is positioned to interact with Nrf2 residue E79. For clarity, the white sticks show only the core ETGE motif from the bound 16-mer peptide. (D) In the presence of small molecule ligands, the R415 side-chain (green) can adopt a position that points out of the binding site, opening the central channel for occupancy by part of the compound (yellow sticks).