Abstract
An opportunity to improve cancer outcomes with machine learning.
Tumors change in response to therapies, but why and how these changes lead to therapy resistance is not well understood, especially when the changes involve the surrounding tissues and conditions called the tumor microenvironment (TME). Powerful machine learning techniques to decode the complex interplay of tumors and the effects of anticancer therapies (e.g., chemotherapy) promise to transform our understanding of therapy-induced tumor remodeling, to develop new targetable pathways for therapy maintenance, and to improve patient outcomes.
By coupling machine learning techniques with new ways to measure the TME at the biomolecular level and changes in clinical cancer management, we will markedly strengthen our understanding of the role that the TME plays in directing tumor response to therapies. New biomolecule measurement technologies, such as multispectral immunohistochemistry (IHC), and high-quality gene expression data from archived tissues will enable deeper views into TME biology. Changes to clinical cancer management, such as the use of neoadjuvant chemotherapy, allow data to be collected from the tumor compartment before and after therapy. Advanced machine learning tools will sift new and biologically important “signals” out of these high-dimensional datasets.
SEEKING OUT FUTURE TUMOR RESPONSE
Clinical perturbations, such as chemo- or radiotherapy, are often based on a simple model of biological causality. For example, a chemotherapy-sensitive tumor will shrink in proportion to the intensity of the chemotherapy it receives. However, the dynamics of complex biological systems are rarely truly linear. Instead, a tumor’s response to therapy can induce remodeling of the TME that drives future responses, including disease progression or therapy resistance. Circumventing or managing these adverse responses requires a systematic view of the interaction dynamics of a tumor, its microenvironment, and clinical perturbations. Despite some progress in this direction, the TME’s great therapeutic and scientific potential remains unrealized because of two limiting factors.
RATE-LIMITING FACTORS
First, the datasets we can now produce using tumor sequencing and imaging technologies are expansive and high-dimensional, a richness that tends to confound current linear thought and analyses. For example, integrating transcriptomic and multispectral IHC data for a pair of patient-matched tumors pre- and post-chemotherapy—at only two time points—presents millions of potential interactions among the transcriptional changes of hundreds to thousands of genes, in addition to composition and spatial information on the microenvironment dynamics of multiple cellular markers. The sheer breadth and complexity of such data has so far inhibited their full use in identifying biologically meaningful and actionable differences. Second, the TME and its response are likely patient and cancer specific. Despite these challenges, we now have newer technology to aid in understanding the nuanced underlying dynamics of the TME.
UNDERSTANDING THE DATA
Modern machine learning techniques provide the foundation to solve these issues, allowing us to simplify complex datasets and to identify biologically meaningful hypotheses about the TME’s response kinetics and its relationship to disease progression (Fig. 1). Unsupervised machine learning and network analysis are powerful tools for reducing the dimensionality and complexity of these datasets so that researchers can explore them to generate new hypotheses. Supervised techniques can then be used to convert high-dimensional measurements into accurate predictions of outcomes or sharp tests of particular mechanistic hypotheses. However, the ultimate utility of these techniques depends critically on the quality and integrity of their data and on the clinical validation of the patterns and predictions they produce.
Fig. 1. Decoding cancer therapy–induced remodeling via machine learning.
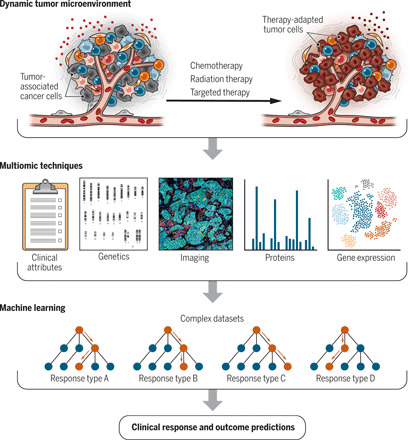
Anticancer therapy significantly remodels the tumor microenvironment (TME). The extent of remodeling can be quantified through multiple biomolecular techniques. Applying machine learning and network analysis to clinical data and complex multiomic datasets has the potential to reshape our ability to anticipate clinical and therapeutic outcomes. Credit: Kellie Holoski/Science Advances.
The recently announced Tumor Profiler Study (1) is an exciting new effort in this direction, with substantial potential to advance our understanding of the TME’s role in disease. Three aspects of its design make it a model study for the TME: (i) integrating machine learning methods with detailed multiomic TME and kinetic data for a large and diverse population; (ii) developing a sustainable model of the translatability of costs, technical expertise, assay limitations, data platform integration, and required infrastructure; and (iii) using profiling and machine learning results to change clinical practice by identifying patient- and tumor-specific vulnerabilities, i.e., precision medicine, through complex datasets.
UNTANGLING TUMOR REMODELING
More concretely, two recent studies—one on pancreatic cancer and one on ovarian cancer, both among the deadliest of human cancers—demonstrate the vast potential of new measurement platforms, and the new insights produced by straightforward machine learning techniques suggest vast untapped potential for more advanced techniques to untangle the underlying dynamics of therapy-induced remodeling of the TME.
A key challenge with treating pancreatic cancers is their general unresponsiveness to immunotherapy. Farren and colleagues (2) used transcriptomic and proteomic analysis to study the effect of surgery, chemotherapy, or chemotherapy plus radiotherapy on the TME of pancreatic cancer. Farren et al. found that radiotherapy treatment profoundly reshapes the TME’s immune characteristics—seen in changes to both transcripts and proteins—while chemotherapy and surgery treatments do not. In particular, tumors treated with radiotherapy showed significant decreases in immune-suppressive factors, including regulatory T cells and an immune checkpoint receptor, which suggests that the immunoresistance of pancreatic tumors may be mediated by the TME. The team also found promising leads for identifying the molecular mechanisms of that connection: therapy-induced expression changes in the RUNX1 and SND1 transcription factors. Although the functional roles of these factors in pancreatic cancer progression and therapeutic response are yet unknown, multiomic techniques indicate an association between RUNX1 and SND1 and critical regulatory nodes.
In our own work on advanced ovarian cancer, we used multiomic techniques to characterize the ovarian TME before and after chemotherapy in patients (3), thereby measuring individual therapy-induced changes. Unlike pancreatic cancer chemotherapy, ovarian cancer chemotherapy had a profound but variable remodeling effect on the TME. Across patient-matched samples, we found significant treatment-induced expression of proinflammatory cytokines, which is consistent with past results. Protein analysis indicated that chemotherapy “activated” pro-oncogenic signaling pathways such as MAPK and JAK/STAT. These findings suggest that chemotherapy alters the TME in a way that drives subsequent disease recurrence and response to second-line therapies. Analyzing the network of transcription factors generated two specific leads for molecular mechanisms that underpin this connection: the activation of the CEBP/β-transcriptional program and the induction of a target gene IER3. As in the pancreatic cancer study, simple machine learning and network analysis identified RUNX1 and SND3, much as they helped identify the novel role of CEBP/β and IER3 in ovarian cancer TME remodeling. The function of these factors in reshaping the TME is largely unknown but provides a valuable direction for future investigation.
TARGETING DIFFICULT CANCERS
This pair of studies are early examples of the potentially transformational utility of combining new measurement and gene transcript technologies with patient-matched pre- and post-treatment tumor samples and using advanced machine learning techniques to uncover new and possibly targetable biology in difficult-to-treat cancers. However, far more can and must be done to exploit the enormous richness and complexity of such multiomic pre- and post-therapy datasets. Decoding the dynamic TME will require a full embrace of machine learning and networks analysis techniques, which we believe will ultimately transform our understanding of tumor biology, therapeutic response, and precision medicine.
REFERENCES
- 1.Irmisch A., Bonilla X., Chevrier S., Lehmann K.-V., Singer F., Toussaint N. C., Esposito C., Mena J., Milani E. S., Casanova R., Stekhoven D. J., Wegmann R., Jacob F., Sobottka B., Goetze S., Kuipers J., Del Castillo J. S., Prummer M., Tuncel M. A., Menzel U., Jacobs A., Engler S., Sivapatham S., Frei A. L., Gut G., Ficek J., Miglino N., Consortium T. P., Aebersold R., Bacac M., Beerenwinkel N., Beisel C., Bodenmiller B., Dummer R., Heinzelmann-Schwarz V., Koelzer V. H., Manz M. G., Moch H., Pelkmans L., Snijder B., Theocharides A. P. A., Tolnay M., Wicki A., Wollscheid B., Rätsch G., Levesque M. P., The Tumor Profiler Study: Integrated, multi-omic, functional tumor profiling for clinical decision support. Cancer Cell 39, 288–293 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Farren M. R., Sayegh L., Ware M. B., Chen H.-R., Gong J., Liang Y., Krasinskas A., Maithel S. K., Zaidi M., Sarmiento J. M., Kooby D., Patel P., El-Rayes B., Shaib W., Lesinski G. B., Immunologic alterations in the pancreatic cancer microenvironment of patients treated with neoadjuvant chemotherapy and radiotherapy. JCI Insight 5, e13036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordan K. R., Sikora M. J., Slansky J. E., Minic A., Richer J. K., Moroney M. R., Hu J., Wolsky R. J., Watson Z. L., Yamamoto T. M., Costello J. C., Clauset A., Behbakht K., Kumar T. R., Bitler B. G., The capacity of the ovarian cancer tumor microenvironment to integrate inflammation signaling conveys a shorter disease-free interval. Clin. Cancer Res. 26, 6362–6373 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]


