Fig. 2. An rRNA switch is central to CDR folding.
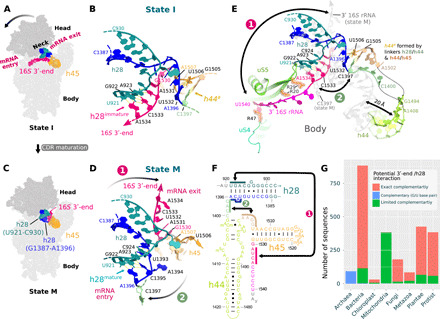
Two conformations of h28 are observed in states I (inactive 30S) (A and B) and M (mature 30S) (C and D) and define an rRNA secondary structure in the CDR. The conformational change is shown schematically (A and C) and in models of the neck region (h28; B and D). The 16S 3′-end swaps between the mRNA entry (B) and exit channels (D; arrow 1) and h28 switches from h28immature (B; U921-A923:U1532-A1534) to h28mature (D; U921-A923:U1393-A1396). In state I (B), U1393-A1396 and subsequent residues in the h28/44 linker (green) can interact with the h44/45 linker (brown) to form a labile helix-like h44a (poorly defined in the cryo-EM map; see fig. S8C). (E) Superimposed CDR structures in states I (colored) and M (gray) highlight changes outside the h28 region, for example, a displacement of h44. The CDR in all states is shown in fig. S7, with supporting cryo-EM maps in fig. S8. (F) Residues involved in the rRNA secondary structure switch are indicated by solid-colored bars where arrow 1 indicates base pairing in h28immature and arrow 2 the formation of h44a. (G) Bar graph comparing the number of 16S rRNA sequences, in various phylogenetic groups, with exact complementarity between the 16S 3′-end (U1532 to A1534) and h28 (residues U921 to A923; pink barks) and those with limited complementarity (≥1 mismatches; green bars). Archaea sequences are complementary in these regions but are colored distinctly to reflect the fact that the complementarity is formed by a non–Watson-Crick, G923:U1532, base pair (41).
