Fig. 5. Sleep defects upon glial kismet loss are caused by developmental hyperserotonemia.
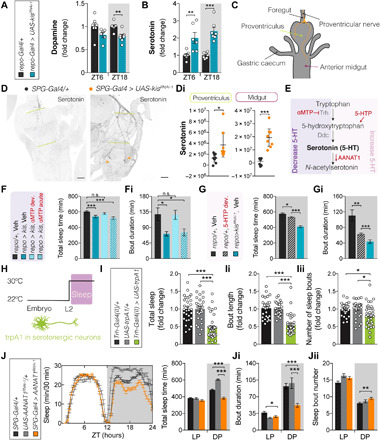
(A and B) repo-Gal4,UAS-Dcr2 > UAS-kisRNAi-1 (blue, n = 6 to 7) exhibit decreased dopamine at ZT18 (P = 0.0028) and ≥2-fold increased serotonin at ZT6 (P = 0.0077) and ZT18 (P = 0.0004) compared to controls (black, n = 7). (C) Scheme of the larval digestive tract and serotonergic innervations. (D) Representative images and (Di) quantification of α-serotonin immunolabeling. SPG kismet knockdown (R54C07-Gal4 > UAS-kisRNAi-1, orange, n = 8) causes increased serotonin in distal innervations (arrowheads) and diffuses signal in the proventriculus (P = 0.037) and anterior midgut (P = 0.0002) compared to controls (black, n = 8). Scale bars, 50 μm. (E) Serotonin metabolism in Drosophila and used approaches to manipulating it. (F, G, and J) Sleep profiles and/or quantification. (Fi, Gi, and Ji) Average duration and (Jii) number of sleep bouts during LP (ZT0 to ZT12) and DP (ZT12 to ZT24). (F and Fi) Pan-glial knockdown (repo-Gal4,UAS-Dcr2 > UAS-kisRNAi-1, dark blue, n = 32) fed with αMTP during development (light blue, n = 49), but not adulthood (light blue dotted, n = 30), shows restored TST and sleep bout duration to vehicle-reared controls (black, n = 41). (G and Gi) Increasing serotonin in wild type (hatched black, n = 72) by developmental 5-hydroxytryptophan (5-HTP) feeding leads to decreased sleep due to significantly reduced sleep bout length compared to vehicle-fed animals (black, n = 70), phenocopying kismet knockdown (blue, n = 62). (H) Temperature program for thermogenetically controlled larval sleep recordings: (I to Iii) Thermogenetic activation of serotonergic neurons [Trh-Gal4(II) > UAS-dTrpA1, green, n = 37] leads to decreased sleep amount (P < 0.0001), bout length (P < 0.0001), and number (P = 0.035 and P = 0.02, respectively) compared to parental controls (Trh-Gal4(II)/+, n = 29; UAS-dTripA1/+, n = 28). (J to Jii) SPG AANAT1 knockdown (SPG-Gal4 > UAS-AANAT1RNAi-1, n = 97) leads to decreased total sleep (P < 0.0001) and bout length (P < 0.0001) compared to controls (SPG-Gal4/+, n = 125; UAS-AANAT1RNAi-1/+, n = 37). Data are represented as means ± SEM, except (Di) represented as means ± 95% confidence interval. (A, B, and Di): Two-tailed unpaired Welch’s t test. (F, Fi, G, Gi, H, I, Ii, and Iii): Kruskal-Wallis test with Dunn’s correction. (J, Ji, and Jii): One-way analysis of variance (ANOVA). Bonferroni correction for multiple testing. P values: *P < 0.05, **P < 0.01, and ***P < 0.001. n.s., not significant. See fig. S7.
