Fig. 8.
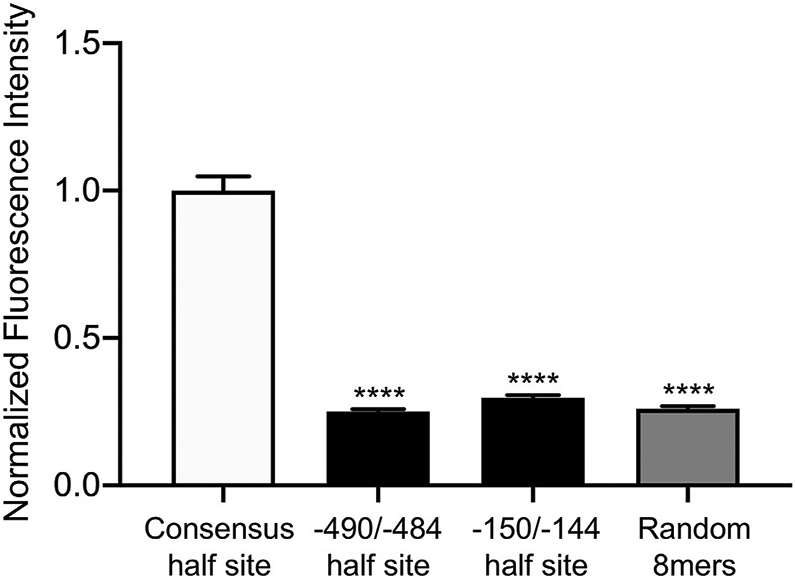
Nonconsensus half sites within the −499/−484 and −159/−144 HREs have decreased affinity for AR compared to the consensus half site. We obtained in vitro binding data for AR from the UniPROBE database (Hume et al., 2015; Mariani et al., 2017). The DBD of AR was tested for its ability to bind every possible 8mer DNA sequence. From this dataset, we extracted the raw fluorescence intensities generated by the AR DBD binding to all 8-mer sequences containing, from left to right, the HRE consensus motif 5′ AGAACA 3′, the −490/−484 nonconsensus half site 5′ TGCTTA 3′, the −150/−144 nonconsensus half site 5′ AGCACT 3′, and randomly-selected 8-mer sequences (n = 48 sequences per category). The raw fluorescence intensity for each sequence was normalized to the average intensity of all 48 sequences containing the HRE consensus motif. Normalized fluorescence values are represented as mean ± SEM for each category of 8-mer sequences listed above. Data were analyzed by one-way ANOVA with Tukey post hoc analysis. Stars indicate significant results compared to consensus half site. ****, p < 0.0001.
