Figure 4. Distinct cellular communities define DLBCL LME categories.
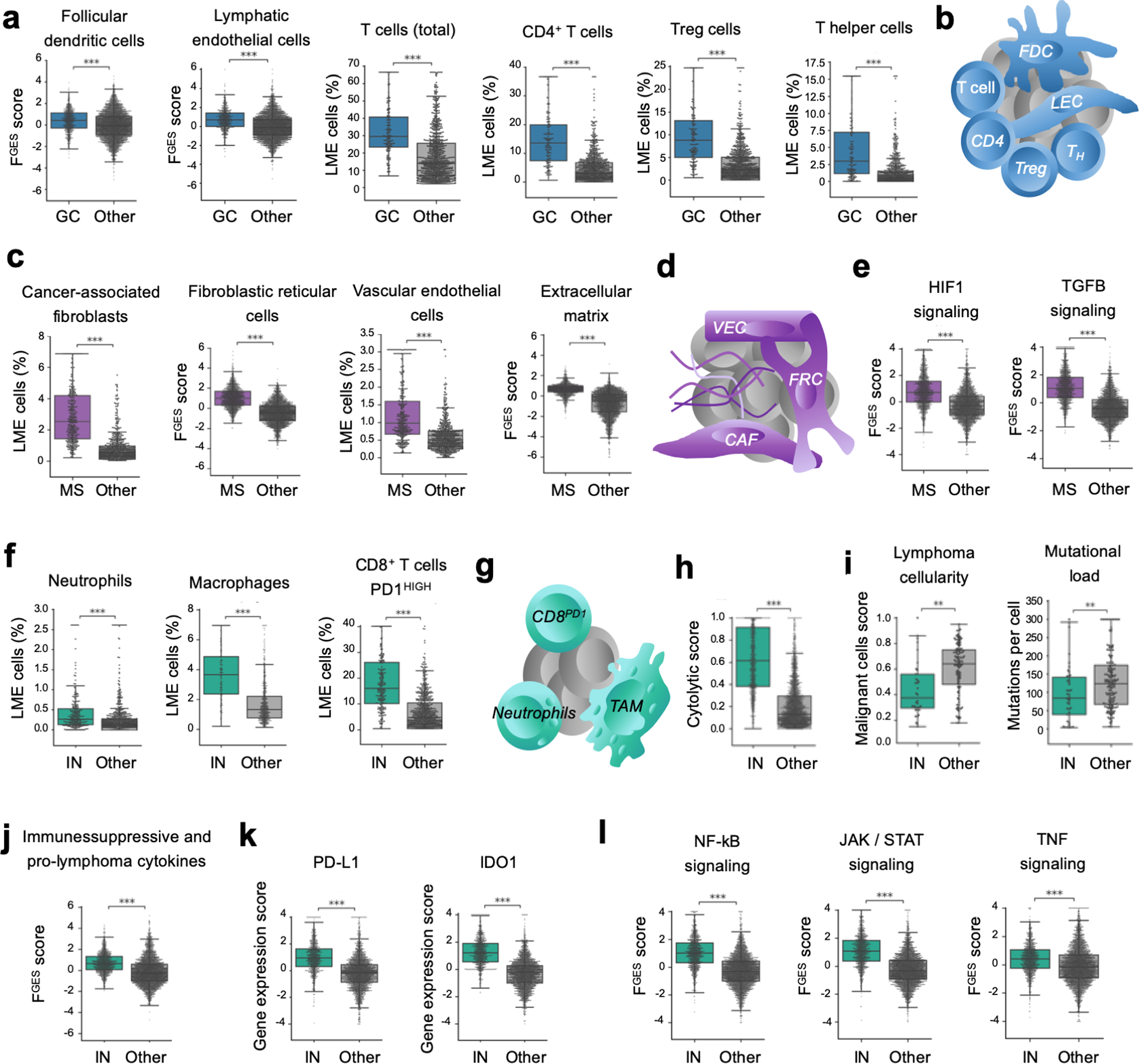
a. Proportion of LME cells significantly enriched in the GC-like LME obtained by cell deconvolution algorithms or functional gene expression signatures. b. Schematic representation of selected features of GC-like-LME. FDC: follicular dendritic cells, LEC: lymphatic endothelial cells. TFH: follicular T helper cells. c. Proportion of LME cells significantly enriched in the MS LME obtained by cell deconvolution algorithms or functional gene expression signatures. d. Schematic representation of selected features of MS-LME. VEC: vascular endothelial cells, FRC: fibroblastic reticular cells, CAF: cancer-associated fibroblasts. e. HIF1 (hypoxia) and TGFB pathways activity. f. Proportion of LME cells significantly enriched in the IN LME obtained by cell deconvolution algorithms. g. Schematic representation of selected features of IN-LME. TAM: tumor-associated macrophages. h. Cytolytic score in the IN-LME vs. other LMEs. i. Lymphoma cellularity (proportion of malignant cells) and mutational load (number of mutations per cell) in DLBCL containing IN-LME vs. other LMEs. j. Immune suppressive / pro-lymphoma cytokines FGES in the IN-LME vs. other LMEs. k. Expression of PD-L1 and IDO1 in lymphomas with IN-LME vs. other LMEs. l. NF-kB, JAK/STAT and TNF-alpha pathways activity. **p<0.01, ***p<0.001
