Abstract
Targeted monotherapies usually fail due to development of resistance by a subgroup of cells that evolve into recurrent tumors. Alveolar rhabdomyosarcoma is an aggressive myogenic soft tissue cancer that is associated with a characteristic PAX3-FOXO1 gene fusion encoding a novel fusion transcription factor. In our myoblast model of PAX3-FOXO1-induced rhabdomyosarcoma, de-induction of PAX3-FOXO1 simulates a targeted therapy that antagonizes the fusion oncoprotein. This simulated therapy results initially in regression of the primary tumors, but PAX3-FOXO1-independent recurrent tumors eventually form after a delay. We report here that upregulation of the fibroblast growth factor FGF8, a direct transcriptional target of PAX3-FOXO1, is a mechanism responsible for PAX3-FOXO1-independent tumor recurrence. As a transcriptional target of PAX3-FOXO1, FGF8 promoted oncogenic activity in PAX3-FOXO1-expressing primary tumors that developed in the myoblast system. In the recurrent tumors forming after PAX3-FOXO1 de-induction, FGF8 expression was necessary and sufficient to induce PAX3-FOXO1-independent tumor growth through an autocrine mechanism. FGF8 was also expressed in human PAX3-FOXO1-expressing rhabdomyosarcoma cell lines and contributed to proliferation and transformation. In a human rhabdomyosarcoma cell line with reduced PAX3-FOXO1 expression, FGF8 upregulation rescued oncogenicity and simulated recurrence after PAX3-FOXO1-targeted therapy. We propose that deregulated expression of a PAX3-FOXO1 transcriptional target can generate resistance to therapy directed against this oncogenic transcription factor and postulate that this resistance mechanism may ultimately be countered by therapeutic approaches that antagonize the corresponding downstream pathways.
Keywords: Targeted therapy, FGF8, PAX3-FOXO1, tumor recurrence
Introduction
Alveolar rhabdomyosarcoma (ARMS) is an aggressive pediatric soft tissue sarcoma that occurs frequently in adolescents and young adults and is characterized by early metastasis, poor response to chemotherapy, and frequent relapses after chemotherapy (1). This cancer usually occurs within skeletal muscle and is hypothesized to develop from skeletal muscle progenitors (2). ARMS is distinguished by a recurrent 2;13 chromosome translocation involving the PAX3 and FOXO1 genes and less frequently by a 1;13 translocation involving the PAX7 and FOXO1 genes. These translocations generate PAX3-FOXO1 (P3F) or PAX7-FOXO1 fusion genes (3,4), which encode novel chimeric transcription factors.
We previously demonstrated that expression of P3F, along with MYCN, induces transforming and tumorigenic activity in human myoblasts, and that P3F plays a crucial role in regulating the proliferation, survival and differentiation of these cells (5). These properties make P3F an interesting therapeutic target in the treatment of ARMS, particularly given the fact that conventional treatment, combining chemotherapy, surgery and radiotherapy, is often ineffective, with a 5-year overall survival less than 50%. Though efforts to develop effective P3F-directed targeted therapy are ongoing, it is well known that resistance to a single therapeutic agent often develops and thus may ultimately occur in monotherapies targeting the P3F oncoprotein. We propose that a model of P3F targeted therapy will be useful in the investigation of such resistance mechanisms. To simulate this targeted therapy, we designed a doxycycline-inducible P3F (iP3F) expression construct to control the timing and level of P3F expression in vitro and in vivo (6). We observed that human Dbt myoblasts engineered with constitutive MYCN and iP3F expression constructs (Dbt/MYCN/iP3F) rapidly form primary rhabdomyosarcoma (RMS) tumors when injected into mice fed a doxycycline-containing diet. When doxycycline induction is stopped to turn off P3F expression, tumors initially regress, and then recurrent tumors without P3F expression form after a variable delay. These findings suggest that a subset of tumor cells is able to conserve their tumorigenicity by developing or maintaining mechanisms that lead to clonal expansion and formation of P3F-independent recurrent tumors. We hypothesize that P3F is required for initial tumor development but is not needed in later stages of tumorigenesis due to the accumulation of additional oncogenic changes.
In this study, we analyzed the expression profile of primary and recurrent tumor-derived (TD) cells and found that fibroblast growth factor 8 (FGF8), a transcriptional target of P3F, often remains upregulated in recurrent TD cells. Furthermore, assays of oncogenic activity reveal that FGF8 is required for proliferation, transformation, migration and invasion of these recurrent TD cells. In addition, FGF8 is also necessary for the oncogenic transformation of primary TD cells as well as human ARMS tumor cells, and FGF8 overexpression in human ARMS cells is sufficient to maintain their oncogenicity when P3F expression is repressed. Our findings provide biological information regarding the consequences of therapeutically targeting oncogenic fusion proteins and may enable development of combined approaches to effectively treat these aggressive cancers.
Materials and Methods
Cell culture
Engineered derivatives of a Duchenne muscular dystrophy myoblast cell line (Dbt) and human RMS cell lines were cultured as described previously (5,7). The source of the cell line is as follow: Dbt, Dr D. Trono; RH30 American Type Culture Collection; Rh28 Dr. B. Emanuel; Rh5 Dr. J. Khan; CW9019 Dr. J. Biegel; Rh41 Dr. C. Linardic; MP4 Dr. T. Cripe. Verification of cell line identity was performed using the short tandem repeat genotyping analysis with the AmpFLSTR profiler plus polymerase chain reaction amplification kit (Applied Biosystems, Foster City, CA). Cell lines were periodically checked with a PCR-based mycoplasma detection kit (ATCC, # 30–1012K, Manassas, VA) to rule out the possibility of mycoplasma contamination.
Transfection and lentiviral transduction
Dbt cells were previously transduced either with a constitutive MYCN expression construct (Dbt/MYCN) or with MYCN and an inducible P3F expression construct (Dbt/MYCN/iP3F) (5,6). The pCMV6-Entry vector expressing FGF8b cDNA or the empty vector EV (Origene, Rockville, MD) was stably transfected using lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA). CRISPR/Cas9 targeting of FGF8 was performed with single-guide RNAs cloned into the lentiCRISPR v2-eGFP (GenScript, Piscataway NJ) and compared with the control non-targeting vector lentiCRISPR v2-eGFP (CR-control). Dbt/MYCN/iP3F cells were transduced with a green fluorescent protein (GFP) expression construct as previously described (6). Lentiviral particles were packaged using HEK293T cells with the human immunodeficiency virus-based pPACK-H1 Packaging Plasmid Mix (System Biosciences, Mountain View, CA).
RNA extraction and gene expression profiling
Total RNA was extracted using the AllPrep Kit (Qiagen, Germantown, MD). Synthesized cDNA was hybridized to the GeneChip™ Human Transcriptome Array 2.0 (#902162, Affymetrix, Santa Clara, CA). Generated CEL files were analyzed using Partek® Flow® software, version 8.0.19.111© (Partek, St. Louis, MO), using the standard recommendations provided by the software. Gene expression changes with >1.5- or <−1.5-fold alterations and FDR (Benjamini and Hochberg-adjusted P-value) <0.01 were considered significant. GSEA analysis was performed using online tools (www.gsea-msigdb.org). Taqman gene expression assays (Life Technologies, Carlsbad, CA) were used to quantify expression of PAX3-FOXO1 (Hs03024825), FGF8 (Hs00171832_m1), PCBD1 (Hs01548084_g1), DUSP6 (Hs04329643_s1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Hs02758991). The 2−Δ Δ Ct method (8) was used to assess the gene expression change.
The data presented in the current publication have been deposited in and are available from the dbGaP database under dbGaP accession phs002344.v1.p1
Protein extraction and western blotting
Protein extraction and western blotting were performed as described previously (9). Membranes were incubated overnight with antibodies against FOXO1 (1:1000, no. 2880; Cell Signaling Technology, Danvers, MA), FGF8 (1:1000, no 16124, Sino Biological, Wayne, PA) and GAPDH (1:1000, no. sc-25778; Santa Cruz Biotechnology, Santa Cruz, CA).
Proliferation, focus formation and clonogenic assays
To monitor proliferation,1000 cells were seeded per well in 96-well plates and imaged in the IncuCyte S3 Live-Cell Analysis System (Essen BioScience, Ann Arbor, MI) every 6 hours for up to 7 days. To assay in vitro oncogenic transformation, 360 transduced Dbt cells were mixed with 360,000 NIH 3T3 cells (1/1000 ratio). Cells were seeded into 60 mm dishes containing a 50:50 mixture of F-10 and DMEM medium, which was renewed biweekly. Clonogenicity was measured in a colony assay by plating 360 Dbt cells in 60 mm plates with F-10 medium. For focus and colony assays, the medium was removed after 2 weeks, and the cells were fixed using methanol and stained with Giemsa. Images were taken using the ChemiDoc XRS+ Imager (BioRad, Hercules, CA). For each condition, foci were counted in three biological replicates using Image J software (10).
Conditioned medium (CM) preparation
After growing cells to 80% confluency, plates were washed 3 times with PBS before adding serum-free growth media to culture the cells for another 24 hours. CM was collected and centrifuged at 1000 rpm for 5 minutes to remove floating cells.
Migration and invasion assays
Migration and invasion assays were conducted using the 24-well format Cell Migration Assay Kit and ECM Matrix Cell Invasion Assay Kit (Sigma-Aldrich, St. Louis, MO). For the invasion assay, transwells with basement membranes were incubated in F-10 serum free media for at least 2 hours prior to the experiment. Cells in serum-free F-10 medium were seeded into the upper chamber at 5 × 104 per well for migration assays and at 2 × 105 per well for invasion assays, while F-10 medium with 15% FBS was added into the lower chamber as chemoattractant. To prevent a difference in the proliferation rate between control and knockout cells, hydroxyurea (Sigma-Aldrich) was added to both chambers, at the indicated concentrations, throughout the experiment.
Immunodepletion assay
CM was concentrated to a volume of 1 ml using the Amicon® Ultra-4 Centrifugal Filter Unit (Millipore, Burlington, MA) and the flow-through was stored at 4°C. The concentrated CM was incubated with FGF8 antibody or IgG control antibody (Cell Signaling Technology) for 1 hour at 4°C, and then incubated overnight with Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology). The protein-antibody-bead complexes were pelleted, washed and resuspended in Laemmli buffer and fractionated by SDS-PAGE electrophoresis. The immunodepleted CM was mixed with the flow-through described above and used for focus assay experiments.
In vivo oncogenic assay
All experimental procedures were approved by the NIH Animal Care and Use Committee. Dbt myoblasts (106 cells) suspended in 50 μl of HBSS (Gibco, Grand Island, NY) were injected into the left gastrocnemius muscle of female 4–6-week-old female NOD-SCID mice (strain #560) (Charles Rivers Laboratories, Wilmington, MA). Mice were fed a regular diet (Harlan Laboratories, Indianapolis, IN), which was replenished two times per week. The mice were observed for tumor formation two times per week for 90 days. Tumor volume was measured as described previously (6). At the time of euthanization, tumors were excised, and cell lines were generated as described (5).
Statistical analysis and data presentation
In vitro experiments were performed in three biological replicates and representative results are shown in the figures. Results are presented as mean ± SE (Standard Error). Quantitative data were analyzed for significance using a two-sided unpaired Student’s t-test as indicated in the figure legends. Results were deemed to be statistically significant when P≤0.05. Prism 8.4.3 software (GraphPad Software, San Diego, CA, USA) was used for data presentation and statistical analysis.
Results
Recurrent tumors retain few upregulated P3F targets
To analyze the differentially expressed genes associated with P3F-independent tumor recurrence in our myoblast model, we generated cell lines from the primary and recurrent tumors and confirmed the presence of P3F expression in the primary TD cells and the absence of P3F expression in the recurrent TD cells at the mRNA and protein levels. To separate P3F-independent from any P3F-dependent cells in the recurrent tumors, we generated subclones by limiting dilution from 3 recurrent TD cell lines to obtain 7 homogeneous populations (termed 1a-c, 2a-b, 3a-b) with P3F-independent transformation (Supplementary Figure 1A–B). Our analysis of microarray data revealed substantial differences between expression profiles of recurrent TD subclones and primary TD cell lines (Figure 1A and Supplementary Figure 2A). The untreated recurrent TD cells showed 1837 significantly upregulated genes compared to untreated parental Dbt/MYCN/iP3F myoblasts, whereas the doxycycline-treated primary TD cells showed 692 upregulated genes when compared to the same untreated parental cells (Figure 1B, Supplementary Table 1). Pathway and GSEA analyses showed an overlap of differentially expressed genes in both primary and recurrent TD cells with growth factor- and KRAS-related transcriptional signatures (Supplementary Figure 2B).
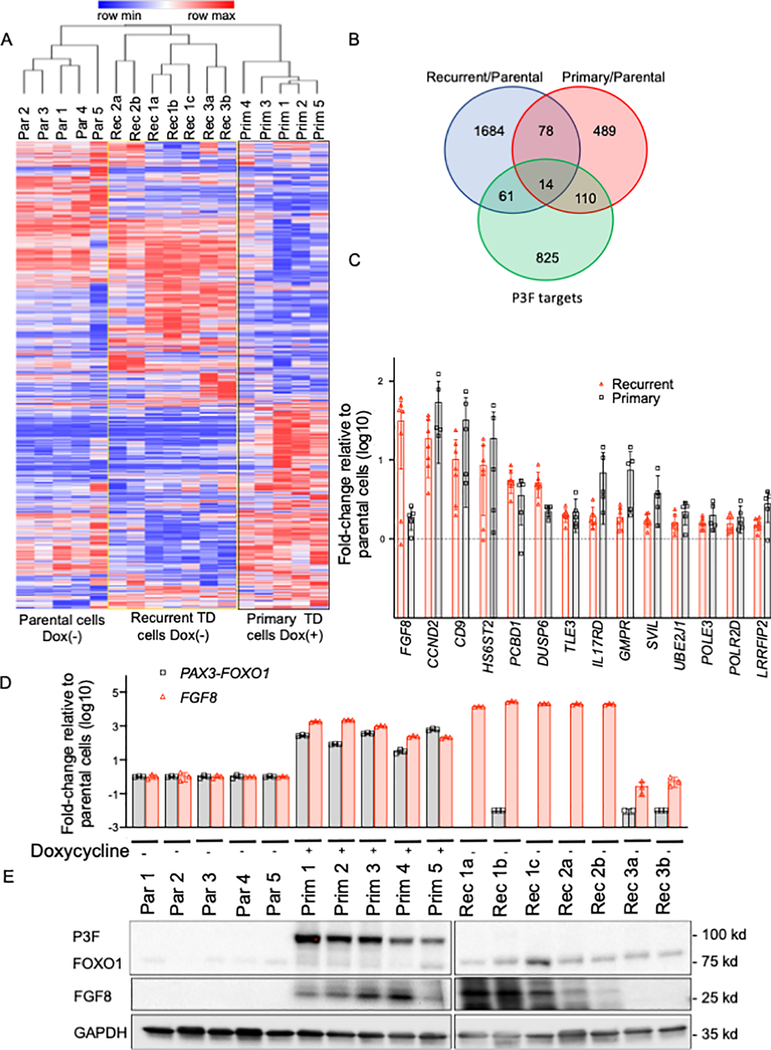
Figure 1. Differences in expression between recurrent and primary TD cells.
A. Hierarchical clustering shows differentially expressed genes between unstimulated parental (Par) cells and recurrent (Rec) TD cells and doxycycline-treated primary (Prim) TD cells. Each column represents an individual cell line (indicated by number) and/or subclone (indicated by letter). The blue and red lines in the heat map indicate low and high gene expression levels according to the scale shown at the top of the heat map. B. Venn diagram showing overlap of the upregulated genes in recurrent TD cells, doxycycline-treated primary TD cells and P3F target genes (11). Differentially upregulated genes were selected using a false discovery adjusted p-value of 0.01 and a 1.5-fold change cut off. C. Analysis of the 14 P3F target genes (described in B) that are significantly upregulated in both recurrent and primary TD cells compared to parental cells. For both primary and recurrent TD cells, array data was used to calculate the expression increase relative to parental cells. Genes are ranked based on the expression increase in recurrent TD cells. P3F (gray bars) and FGF8 (red bars). D. P3F and FGF8 mRNA expression levels in parental cells, primary TD cells and recurrent TD cells. Some cells were treated with doxycycline, as shown. P3F and FGF8 mRNA expression was assayed by real-time RT-PCR and normalized for GAPDH expression by the ΔΔCt method. The data are expressed as the mean +/− SE of 3 replicates. E. P3F and FGF8 protein expression in the three groups of cells (as described in part D). Western blot analysis was performed using GAPDH expression as a control for protein loading.
To search for common features between the primary and recurrent TD cells, comparison of differentially expressed genes (relative to the parental cells) revealed that recurrent and primary TD cells shared 92 upregulated genes, and 14 of these genes are P3F transcriptional targets based on CHiP-seq studies (11) (Figure 1B). Among these 14 P3F target genes, FGF8 showed the highest level of upregulation in recurrent TD cells (Figure 1C). Furthermore, based on the microarray finding that FGF8, PCBD1 and DUSP6 were expressed at higher levels in recurrent TD cells compared to primary TD cells (Figure 1C and Supplementary Figure 3A), we used qPCR assays and confirmed that FGF8 expression was significantly higher in the recurrent compared to primary TD cells (Figure 1D, Supplementary Figure 3B). However, qPCR did not confirm a significant difference in either PCBD1 or DUSP6 expression between primary and recurrent TD cells, even though both are significantly upregulated in primary and recurrent TD cells compared to parental cells (Figure 1C, Supplementary Figure 3C–D). We postulate that continued upregulation of a small number of P3F transcriptional targets may result from additional molecular changes occurring in rare cells in the primary tumors, and the maintained expression of these P3F targets in recurrent TD cells suggests that some P3F targets may be needed in the absence of P3F to maintain oncogenicity and induce recurrent tumor development.
FGF8 knockout impairs recurrent tumor cell oncogenicity
Based on previous findings of FGF8 involvement in colorectal, breast and prostate cancer progression (12–15), we investigated the role of FGF8 in P3F-independent oncogenesis. To study FGF8 involvement in oncogenicity of recurrent tumors, we first confirmed its overexpression in recurrent TD cells both at the mRNA and protein levels. Results from the array analysis indicated that FGF8 is highly expressed (>30-fold) in most of the recurrent TD subclones selected for microarray analysis, compared to the parental unstimulated cells, and this finding was confirmed by qPCR and western blot assays (Figure 1D–E). However, while the array data showed that FGF8 mRNA was only 1.8-fold higher in primary TD cells compared to parental cells, our subsequent experiments showed higher FGF8 expression in primary TD cells at both the mRNA and protein levels. In contrast, FGF8 mRNA and protein expression was not detected in unstimulated parental cells and in two recurrent TD subclones (Rec 3a and Rec 3b).
We utilized a CRISPR-Cas9 (CR) loss of function strategy to knock-out FGF8 in recurrent TD cells with high FGF8 expression. Using 2 different recurrent populations (Rec 1a and Rec 2a), Cas9/gRNA-transduction without selection showed lower FGF8 protein expression with 3 different gRNAs targeting FGF8 (CR-FGF8 #1, FGF8 #2 and FGF8 #3) compared to the non-target control (Figure 2A and Supplementary Figure 4A). To distinguish cells within this transduced Rec 1a population in which FGF8 protein expression was successfully knocked out, we identified subclones that completely lack FGF8 protein expression. Expression profile analysis of the CR-FGF8 subclones lacking FGF8 expression compared to the CR-control subclones showed differentially expressed genes similar to a KRAS-related transcriptional signature (Supplementary Figure 2B).
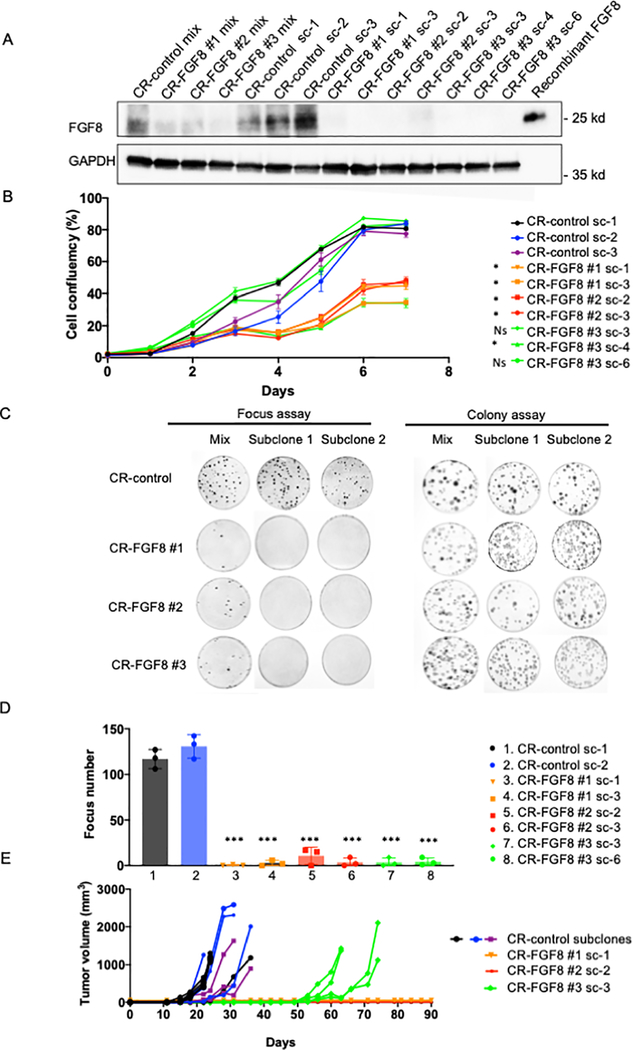
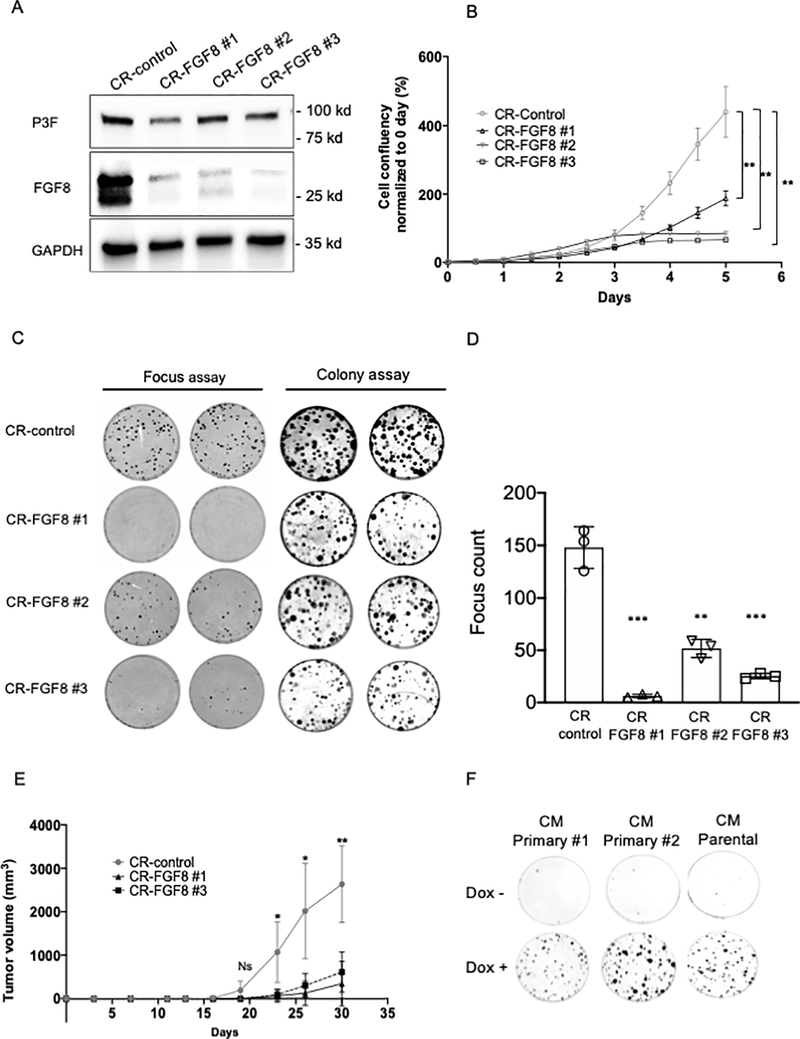
Figure 2. FGF8 knockout impairs transformation and tumorigenicity of recurrent TD cells.
A. FGF8 protein expression in knockout and control cells. FGF8 was knocked-out in Rec 1a cells using lentiviruses containing three different CRISPR constructs: CR-FGF8 #1, CR-FGF8 #2 or CR-FGF8 #3. For a CRISPR control (CR-control), cells were also transduced with the non-targeting construct pLentiCRISPR v2. Western blot analysis was performed to measure FGF8 protein expression in the original 4 transduced populations (mix) and in subclones (sc) derived from these populations. GAPDH was used as a loading control. B. Cell growth of knockout and control subclones from Rec 1a cells was monitored by assessing cell confluency over 7 days using the IncuCyte system. The data are displayed as the mean +/− SE of 4 different wells and are representative of 3 independent experiments. For statistical significance, each CR-FGF8 subclone was compared individually to each of the CR-controls (sc-1, sc-2 and sc-3) for the three last time points (5, 6 and 7 days). An unpaired two-sided Student’s t-test was used and showed that all the comparisons where significant except for the CR-FGF8 #3 sc3 and sc6. * P<0.05, Ns: not significant. C. Focus formation and clonogenicity were assayed in knockout and control cells using the original transduced populations (mix) and subclones derived from these populations. D. Foci counting in transformation assay shown in part C. Foci were counted in triplicate experiments using subcloned populations derived from CRISPR control sc1 and sc2, CR-FGF8 #1, CR-FGF8 #2 or CR-FGF8 #3. Two-sided unpaired Student’s t-test was used to compare each CR-FGF8 subclone to each of the CR control subclones sc1 and sc2. All the differences were statistically significant with a P<0.001 (***). E. Tumor formation in knockout and control cells. Tumor growth curves in NOD-SCID mice (4 mice per group) injected intramuscularly with subclones derived from transduction of CR-FGF8 or CR-control constructs into Rec 1a cells.
As FGF8 is a growth factor, we first analyzed proliferation in CR-FGF8 and control cells. We observed that cell proliferation was significantly diminished in all CR-FGF8 subclones except for two subclones for which proliferation was comparable to the control subclones (Figure 2B). In the original Cas9/gRNA-transduced populations, a focus formation assay showed lower transformation capability of cells transduced with the three FGF8-specific gRNAs compared to the non-target control, and the CR-FGF8 subclones lacking FGF8 expression showed a complete loss of transformation compared to non-targeting control subclones (Figure 2C–D, Supplementary Figure 4B). Of note, subclones from the original CR-FGF8-transduced population that still express FGF8 maintained transformation capability in vitro (Rec 1a/CR-FGF8 #1 sc-2; Supplementary Figure 4, C–D). In contrast to these focus formation results, a clonogenicity assay did not show differences between CR-FGF8 and control cells, either in the original transduced population or the subclones (colony assay, Figure 2C). This finding suggests that FGF8 is necessary for transformation but not clonogenic formation in these recurrent TD cells.
To assess the involvement of FGF8 in recurrent tumor formation in vivo, we performed intramuscular injection of one subcloned population from each of the three FGF8-specific gRNAs in addition to three control subclones into NOD-SCID mice fed a standard diet without doxycycline. We observed that the control subclones started to form tumors 12 days after injection, whereas tumor formation was delayed to 50 days after injection for the subclone derived from CR-FGF8 #3 cells and no tumor was observed for the subclones derived from CR-FGF8 #1 and CR-FGF8 #2 cells (Figure 2E). Furthermore, in a focus assay experiment, we observed that induction of P3F expression in recurrent TD cells (Rec 1a) did not substantially revert the loss of transformation following FGF8 knockout (CR-FGF8 #2, #3) whereas focus formation was enhanced in the CR-control subclone (Supplementary Figure 4E). These combined results suggest that FGF8 is necessary for tumorigenesis after P3F loss in many of the recurrent tumors forming in our myoblast model of RMS.
Given the fact that invasion and migration are important features in cancer progression, and that FGF8 was previously reported to promote these two processes in other cancer types (13,16), we investigated whether FGF8 is involved in the migration and invasion of the recurrent TD cells. To control for any proliferation-related changes due to FGF8 knockout, we used hydroxyurea in both assays and determined the optimal concentration to equalize the proliferation rate in CR-control sc1 and CR-FGF8 #2-sc3 (Supplementary Figure 5A). Our data indicate that the loss of FGF8 expression in recurrent TD cells (CR-FGF8 #2-sc3) resulted in reduced migration and invasion capability of these cells when compared to control knockout cells CR-control sc1 (Supplementary Figure 5B–C). These findings are consistent with the premise that FGF8 also stimulates migration and invasion in our model of P3F-independent tumor recurrence.
FGF8 induces tumorigenicity in Dbt/MYCN cells
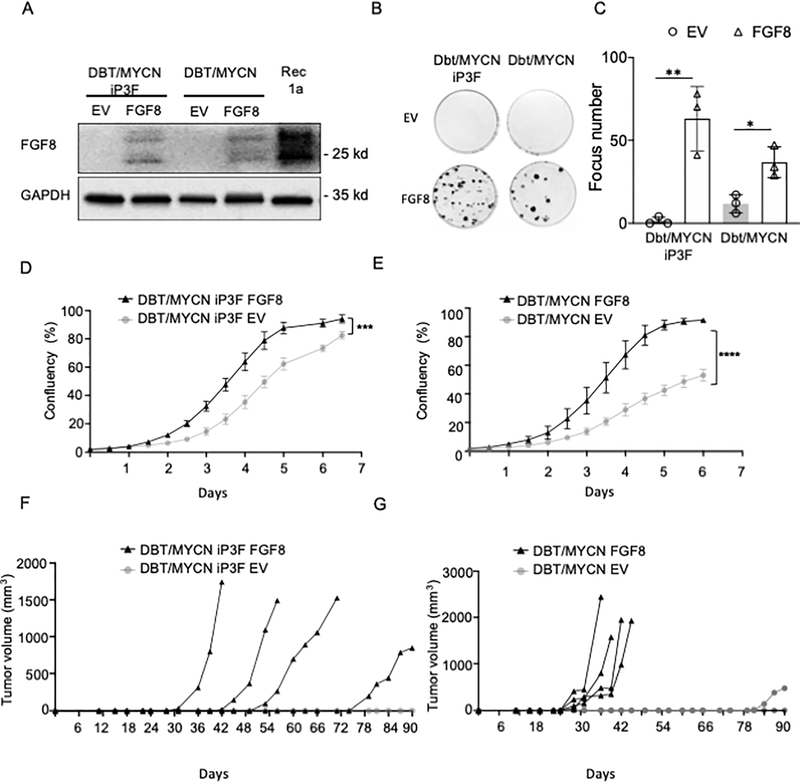
To further investigate whether FGF8 is able to induce tumor recurrence, we overexpressed FGF8 in Dbt/MYCN/iP3F cells and in Dbt/MYCN cells (Figure 3A). In the absence of doxycycline, we detected FGF8 overexpression at lower levels than that found in recurrent TD cells. Expression profile analysis of the Dbt/MYCN/iP3F and Dbt/MYCN cells stably expressing FGF8 relative to the cells with an empty vector (Dbt/MYCN/iP3F-EV Dbt/MYCN-EV) revealed differentially expressed genes that overlap with a KRAS-related transcriptional signature (Supplementary Figure 2B). Furthermore, both cell lines overexpressing FGF8 demonstrated enhanced proliferation rate and focus formation compared to control myoblasts containing the empty vector EV (Figure 3B–E). In addition, to study the effect of FGF8 on tumor growth, Dbt/MYCN/iP3F-FGF8, Dbt/MYCN-FGF8, Dbt/MYCN/iP3F-EV and Dbt/MYCN-EV were injected into NOD-SCID mice. For both cell lines, FGF8 expression promoted tumor formation (Figure 3F–G). Only one small tumor formed from Dbt/MYCN-EV cells; this tumor developed much later than the Dbt/MYCN-FGF8 cells. These results show that FGF8 is able to induce tumor formation, thus indicating that this growth factor is necessary and sufficient for the formation of recurrent tumors in our model.
Figure 3. FGF8 overexpression in Dbt myoblasts induces transformation and tumorigenicity.
A. FGF8 overexpression in Dbt/MYCN/iP3F and Dbt/MYCN cells. FGF8 cDNA or a control construct (EV) was stably transfected into the cells and protein expression was measured by western blotting after 15 days of G418 antibiotic selection (100 μg/ml). GAPDH expression was used as a loading control. B. Representative images of focus formation assay showing oncogenic transformation in both cell lines overexpressing FGF8 but not in cells transfected with the empty vector (EV). C. Foci counting for the transformation assay shown in part B. For each condition, foci were counted in three different plates. The two-sided unpaired Student’s t-test was applied to determine significant differences between the control and test groups. * P<0.05. ** P<0.01. D and E. Growth rate of Dbt/MYCN/iP3F (D) and Dbt/MYCN (E) cells transfected with FGF8 expression construct or EV control construct monitored with the IncuCyte system. The data are displayed as the mean +/− SE of 4 different wells. Two-sided Student’s t-test was used to determine significant changes between the control and test groups. *** P<0.001. F, G. Tumorigenic potential of Dbt/MYCN/iP3F (F) and Dbt/MYCN (G) cells transfected with FGF8 expression construct or the EV control construct. Tumor growth (mm3) was assessed following intramuscular injection of 106 cells into nude mice (4 mice per group). All assays were performed without doxycycline so that P3F was not induced in Dbt/MYCN/iP3F cells.
FGF8 induces cell transformation in an auto/paracrine manner
As a growth factor, FGF8 is secreted into the extracellular space and binds to its membrane receptors to activate downstream pathways. To further investigate FGF8-induced transformation in our model, we collected conditioned medium (CM) from cultures of unstimulated parental cells (Dbt/MYCN/iP3F), Rec 1a recurrent TD cells, Dbt/MYCN-FGF8 cells and Dbt/MYCN-EV cells. In focus formation assays using Dbt/MYCN cells, we observed in vitro transformation only when adding CM from recurrent TD or Dbt/MYCN-FGF8 cells but not with CM from unstimulated parental cells or Dbt/MYCN-EV control cells (Figure 4A, Supplementary Figure 6A). Then, we used western blotting to confirm the presence of FGF8 in the medium from recurrent TD cells but not in the medium from parental cells (Supplementary Figure 6B). In addition, using parental Dbt/MYCN/iP3F cells expressing GFP, we verified that the transformed foci derive from the parental Dbt cells and not from the NIH 3T3 cells (Supplementary Figure 6C). Moreover, in a focus assay with Dbt/MYCN cells, we observed that addition of purified FGF8 into the media induces in vitro transformation in a dose-dependent manner without significant effect on the overall cell clonogenicity (Figure 4B).
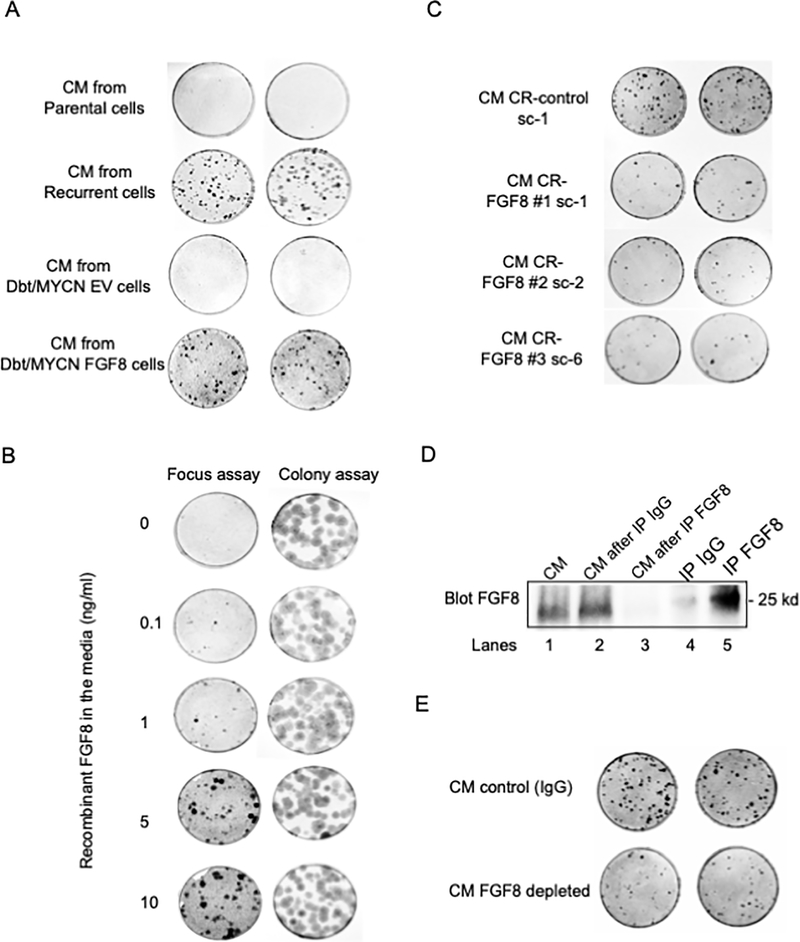
Figure 4. FGF8 promotes cell transformation in an autocrine and/or paracrine manner.
A. Focus formation in Dbt/MYCN cells grown in CM collected from cultures of unstimulated parental Dbt/MYCN/iP3F cells or recurrent TD cells or from Dbt/MYCN cells transfected with FGF8 or EV. The focus assay was performed using Dbt/MYCN cells co-cultured with NIH3T3 cells (Ratio 1/1000) in a medium consisting of 50% Dulbecco’s modified Eagle’s medium and 50% of F-10 CM. B. Focus assay of Dbt/MYCN cells grown in medium supplemented with recombinant FGF8 (R&D Systems) at the indicated concentrations. C. Focus assay of Dbt/MYCN cells grown in CM from the cell culture of the following recurrent TD subclones: CR-control sc-1, CR-FGF8 #1 sc1, CR-FGF8 #2 sc-2 and CR-FGF8 #3 sc-6. D. Western blot studies of FGF8 immunodepletion. Lane 1: FGF8 protein level in CM. Lanes 2–3: FGF8 levels after immunodepleting FGF8 in CM from recurrent TD cell culture compared to treatment of this CM with the IgG control. Lanes 4–5: Immunoprecipitated FGF8 compared to the control IgG. The loading controls are shown in Supplementary Figure 6E. E. Focus assay of Dbt/MYCN cells grown in FGF8-depleted medium compared to the control CM treated with IgG. In parts A, C, and E, two replicates are shown for each CM.
Subsequently, we sought to determine whether removal of FGF8 from the medium will impact cell transformation. First, we used CM from cultures of recurrent TD cells in which FGF8 was knocked-out, as described earlier. In these experiments, CM from CR-FGF8 #1, FGF8 #2, and FGF8 #3 subclones of a recurrent TD line resulted in substantially fewer foci compared to the number of foci formed with CM from CR-control cells (Figure 4C, Supplementary Figure 6D). In a second approach, we performed antibody-mediated depletion of FGF8 from the CM. After collecting antibody-protein complexes with agarose beads, FGF8 depletion in the CM was confirmed by western blot (Figure 4D, Supplementary Figure 6E). In a focus formation assay, FGF8-depleted CM generated a reduced number of foci compared to the control IgG-treated CM (Figure 4E, Supplementary Figure 6F). Together, these results indicate that FGF8 induces transformation of recurrent TD cells in an autocrine/paracrine manner, probably through the activation of FGF receptors on the cell membrane.
FGF8 is involved in the oncogenicity of primary tumor cells
As we found that FGF8, a known P3F target, is upregulated in the presence of P3F in the primary TD cells, we asked whether this growth factor also plays an important role in P3F-induced oncogenicity in primary tumors. When we used a CRISPR-Cas9 approach to knock-out FGF8 in primary TD cells (Figure 5A), we observed that the proliferation rate was reduced using FGF8-specific gRNAs #1, #2 and #3 (Figure 5B). Similarly, the gRNAs #1, #2 and #3 considerably reduced focus formation; this effect on in vitro transformation was greater than the effect on clonogenicity (Figure 5C–D). In addition, cells transduced with the FGF8-specific gRNAs #1 and #3 formed smaller tumors compared to the CR-control tumors (Figure 5E). Additional in vitro studies showed that CM from primary TD cells or parental Dbt/MYCN/iP3F cells induced transformation in cultures of Dbt-MYCN cells only when P3F expression was stimulated with doxycycline (Figure 5F). These results suggest that FGF8 is required for P3F-induced oncogenicity of primary tumors in our myoblast model system.
Figure 5. FGF8 is involved in the oncogenicity of primary tumors from the myoblast system.
A. Western blot analysis of FGF8 expression in doxycycline-treated primary TD cells (Prim 1) after transduction with lentiviruses containing CRISPR constructs targeting FGF8 (#1, 2 or 3) or a non-targeting CRISPR construct (control). P3F induction was confirmed in these cells and GAPDH was used as a loading control. B. Cell growth of doxycycline-treated primary TD cells transduced with CR-FGF8 or CR-control constructs was monitored using the IncuCyte system. The data are displayed as the mean +/− SE of 4 different wells. Statistical significance was determined using unpaired Student t-test. ** P<0.01. C. Representative images of focus formation and clonogenicity that were assayed in doxycycline-treated primary TD cells transduced with CR-FGF8 or CR-control constructs; two replicates are shown for each assay. D. Foci counting from focus formation assay shown in part C. Focus counting was performed in triplicate for the CRISPR groups indicated in part C. The two-sided unpaired Student’s t-test was used to determine significant differences between the control and each test group. ** P<0.01. *** P<0.001. E. Tumor formation using FGF8 knockout and control cells. Tumor growth curves in NOD-SCID mice (5 mice per group) injected intramuscularly with cells from transduction of CR-FGF8 or CR-control constructs into Prim 1 cells. For statistical significance, CR-FGF8 #1 and #3 were compared to the CR-control, for the four last time points. Unpaired two-sided Student’s t-test was used, and significance is indicated for the two comparisons. * P<0.05 ** P<0.01, Ns: not significant. F. Focus assay of Dbt/MYCN cells grown in CM collected from cultures of primary TD cells (Primary #1 or Primary #2) or parental Dbt/MYCN/iP3F cells with (dox+) or without (dox-) doxycycline induction of P3F expression.
FGF8 rescues oncogenicity following loss of P3F expression in human ARMS cells
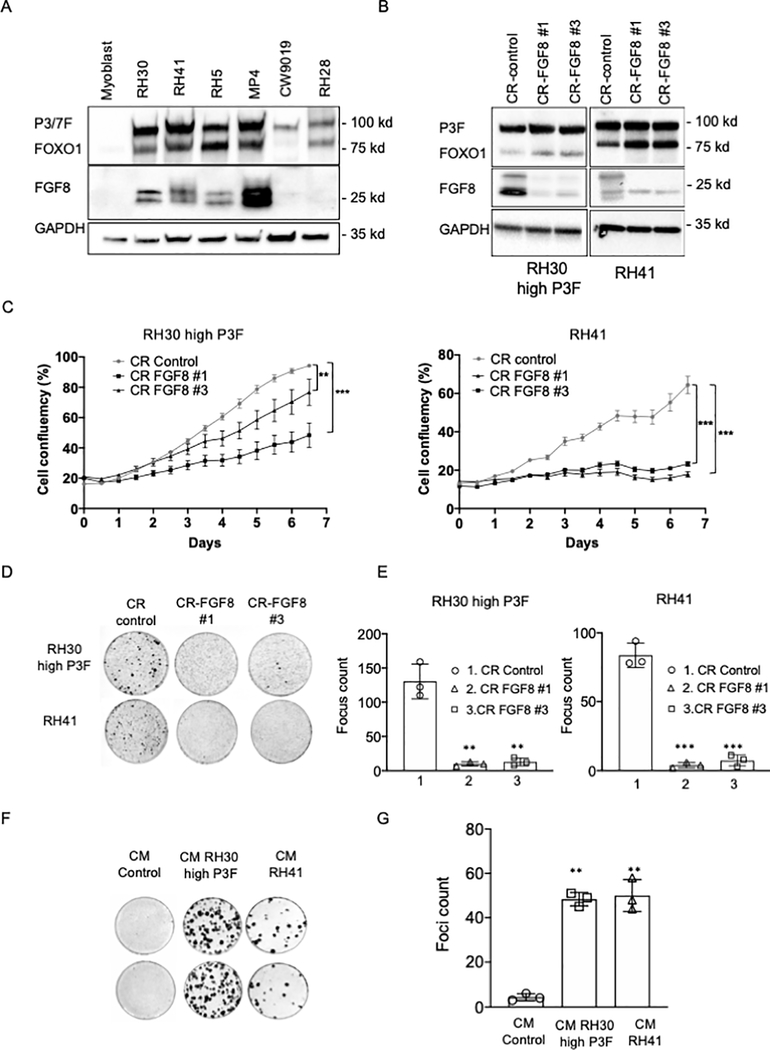
In light of the findings in our myoblast model system, we investigated whether FGF8 contributes to the oncogenicity of human ARMS cells. We analyzed FGF8 expression in a set of ARMS cell lines and observed that FGF8 protein is expressed in the majority of the P3F-positive RMS cell lines but not in the one tested PAX7-FOXO1-positive line (CW9019) (Figure 6A). We then studied FGF8 involvement in the oncogenicity of P3F-positive ARMS cells using RH30 (designated RH30 high P3F) and RH41 cell lines; these cells express both P3F and FGF8 and are transformed in vitro. Using the same CRISPR/Cas9 approach described above, we observed that FGF8 knockout in RH30 and RH41 (Figure 6B) reduced cell growth and significantly impaired cell transformation compared to the CRISPR control (Figure 6C–E). In addition, CM from cultures of RH30 and RH41 cells induced transformation of Dbt/MYCN cells in a focus formation assay (Figure 6F–G). These results confirm a role for FGF8 in the oncogenicity of human FP ARMS cells.
Figure 6. FGF8 involvement in the oncogenicity of human FP RMS cells.
A. Western blot analysis of FGF8 protein expression in a set of human FP RMS cell lines. Fusion status (P3F or P7F) was confirmed by western blot analysis with a FOXO1 antibody, and GAPDH was assessed as a loading control. B. Western blot analysis of FGF8 expression in FP RMS cell lines after transduction of lentiviruses containing CRISPR constructs targeting FGF8 (#1 or #3) or a non-targeting CRISPR construct (control). P3F expression was confirmed by western blot and GAPDH was used as a loading control. C. Cell growth in knockout and control RH30 high P3F (left) and RH41 (right) cells was monitored with the IncuCyte system over a 7-day period. The data are displayed as the mean +/− SE of 4 different wells. Statistical significance between CR control and CR-FGF8 #1 or #3 was determined using unpaired two-sided Student’s t-test. D. Focus formation was assayed in RH30 high P3F and RH41 cells transduced with CRISPR constructs targeting FGF8 or the non-targeting control construct. E. Foci counting for each condition in the transformation assay shown in part D. F. Focus assay performed using Dbt/MYCN cells and CM collected from cultures of Dbt/MYCN cells (CM control), RH30 high P3F and RH41 cells; two replicates are shown for each CM. G. Foci counting corresponding to each condition in part F. For parts C, E and G, the results represent the counting of triplicates, and the two-sided unpaired Student’s t-test was applied to determine significant differences between the control and each test group. ** p < 0.01, *** p < 0.001.
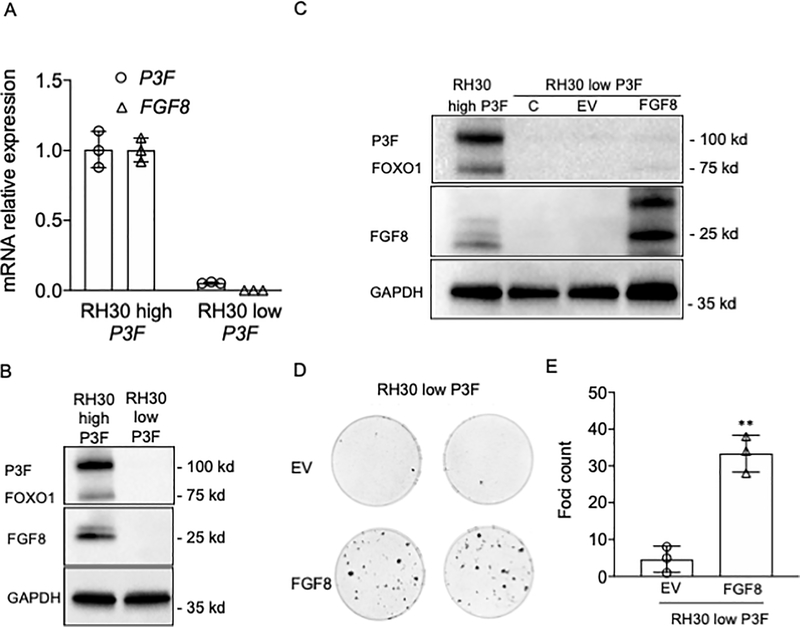
To recapitulate the involvement of FGF8 in a recurrence mechanism in human ARMS cells, we used a spontaneous non-transformed variant of RH30 cells that expresses very low levels of P3F and FGF8 (designated RH30 low P3F) (Figure 7A–B). These non-transformed RH30 low P3F cells are analogous to the primary Dbt/MYCN/iP3F tumors after doxycycline withdrawal. To simulate the recurrence mechanism in our model myoblast system, we stably transfected FGF8 into the RH30 low P3F cells (Figure 7C). These RH30-low-P3F stably expressing FGF8 formed foci in vitro (Figure 7D–E) whereas the RH30-low-P3F transfected with the empty vector did not form foci. These findings demonstrate that FGF8 expression can rescue the loss of P3F expression in these human ARMS cells.
Figure 7. FGF8 rescues transformation after loss of P3F expression in human FP RMS cells.
A. RT-PCR assay of P3F and FGF8 expression in RH30 cells with high or low P3F expression levels. The data are expressed as the mean +/− SE of 3 replicates. B. Western blot assay of P3F and FGF8 protein expression in RH30 cells with high or low P3F expression level. C. FGF8 levels in RH30 low P3F without transfection (C) and after stable transfection with FGF8 expression construct (FGF8) or control construct (EV). D. Focus assay of RH30 low P3F cells stably transfected with FGF8 construct or the control EV construct; two replicates are shown for each assay. E. Counting of foci for each condition in the transformation assay in part D. Results show the number of foci in triplicate experiments of RH30 low P3F stably transfected with EV or with FGF8 constructs. A two-sided unpaired Student’s t-test was applied to determine statistical significance between the control and test groups. ** p < 0.01.
Discussion
Despite continuous advancements in anti-tumor therapies, tumors still frequently recur, months or even years after completing therapy. Targeted therapies may appear to be successful in the short-term but fail due to the development of resistance in a small number of tumor cells and eventual recurrence of the tumor. Resistance to targeted therapy has been observed with a number of approved drugs, such as those targeting ERBB2 in breast cancer or the androgen receptor (AR) in prostate cancer (17–20). Resistance can be caused by molecular alterations that enhance drug efflux (21), prevent drug binding to the target or activate alternative signaling pathways (22,23). Causative molecular alterations include point mutations, gene amplification and other unknown events resulting in increased gene expression (24,25). Therefore, understanding the cellular and molecular events responsible for tumor recurrence is critical to successfully predict and treat these unwanted events.
In this paper, we present evidence for a molecular mechanism underlying a fusion oncoprotein-independent tumor recurrence. In our myoblast system, the de-induction of P3F models a targeted therapy that antagonizes the fusion oncoprotein whereas the recurrence of fusion-independent tumors models acquired resistance to this targeted therapy. Our data reveal that FGF8, a direct transcriptional target of P3F, is upregulated in P3F-independent tumor recurrences. We determined that FGF8 expression promotes tumorigenicity of P3F-positive primary tumors and that upregulation of this growth factor is necessary and sufficient to induce P3F-independent tumor growth through an autocrine mechanism. Therefore, overexpression of FGF8 by P3F-independent events in rare cells is a mechanism that maintains a critical aspect of P3F oncogenicity and permits the development of recurrent tumors in the absence of P3F.
Expression profiling has been a valuable approach to interrogate recurrence mechanisms. In our study, expression profiling revealed biological differences between the primary and recurrent tumors. Such expression pattern differences have also been noted in other studies in which tumors recur following loss of an oncogenic transcription factor or following anti-cancer therapy (25–27). In glioblastomas treated with temozolomide, expression profiling revealed two distinct groups of recurrent tumors, one group with a similar expression profile to matched primary tumors and a second group with reprogramming to a neural-like pattern (28). This observation indicates that more than one mechanism, in this case revealed by expression patterns, can be associated with recurrence of the same tumor type. In our study, several clones isolated from recurrent tumors have P3F-independent transformation capacity but lack FGF8 overexpression, thus indicating the existence of heterogeneity in the mechanism for P3F-independent oncogenesis and tumor recurrence. Further analysis of these and other P3F-independent clones will determine whether different signaling pathways are involved or whether these events can be ascribed to deregulating FGF signaling by other molecular changes such as upregulation of FGF receptors or other FGF family members.
The FGF family includes 23 members, grouped into several subfamilies where the FGF8 subfamily comprises FGF8, FGF17 and FGF18 (29). FGF signaling is deregulated in multiple pathological processes including cancer and impinges on several major downstream signaling pathways (30). High FGF8 expression has been reported in many types of cancers including colorectal, breast and prostate carcinoma (13,31,32), and has been correlated with advanced tumor stage and poor survival (13,33,34). In colorectal cancer, FGF8 expression was associated with tumors that do not respond to neoadjuvant radiotherapy and in vitro experiments showed that FGF8 is upregulated in cancer cells following irradiation or treatment with irinotecan (35,36). In accord with our finding that loss of P3F triggers a survival crisis followed by cell death (6), we postulate that cells overexpressing FGF8 may overcome this survival stress through the activation of downstream signaling pathways that promote survival and proliferation. In colorectal cancer cells, FGF8 overexpression promotes proliferation, clonogenicity and invasiveness in vitro and enhances tumorigenicity in vivo (13). In prostate cancer resistant to AR-directed therapy, the tumorigenicity of AR-independent tumors relies on overexpression of FGF8 (37). Our results in concert with these other findings thus indicate that FGF8-induced tumorigenicity is a frequent mechanism for recurrent tumor development.
The expression of FGF8 is regulated by a number of tissue-specific mechanisms. In prostate cancer, FGF8 is an androgen-induced growth factor (38) that is directly regulated by the androgen receptor through a responsive element located in the FGF8 proximal promoter (39). In fusion-positive (FP) RMS, FGF8 is a direct downstream target of the fusion transcription factor P3F, which binds to a responsive element positioned in the intronic region of the FGF8 gene (11). A recent study demonstrated that the hepatitis BX-interacting oncoprotein (HBXIP) upregulates FGF8 by two mechanisms, activation of its proximal promoter by direct binding to a cAMP response element-binding protein and inhibition of miRNA-503–5p, which directly targets the 3′ untranslated region of FGF8 mRNA (40). In our study, the absence of P3F expression in the recurrent tumors indicates that FGF8 overexpression is maintained at higher levels by an alternative mechanism, which may involve transcriptional or post-transcriptional effects. FGF8 upregulation could be caused by activation of another transcription factor which may be facilitated by the active open chromatin or by a mutational change in one of the regulatory regions of the FGF8 gene. It is also possible that FGF8 upregulation is maintained in the recurrent tumors through increased RNA stability mediated by inhibition of microRNA binding to the 3’UTR, by downregulation of such an inhibitory microRNA or by 3’ UTR shortening. A recent study demonstrated that P3F regulates expression of many microRNAs in FP RMS (41). Of note, the finding that P3F represses miR-503–5p may constitute another pathway by which P3F induces FGF8 expression in primary FP RMS tumors. However, the expected increase in miR-503–5p expression following P3F inactivation would not be responsible for increased FGF8 expression in recurrent tumors without an aberrant change, such as dysregulated miR-503–5p expression or FGF8 3’ UTR shortening. Further elucidation of the mechanism by which FGF8 is upregulated in recurrent tumors, by studies including microRNA profiling, may be useful in developing specific combined targeted therapies.
It should also be noted that DUSP6, another P3F target upregulated in recurrent TD cells, encodes a dual phosphatase that functions as a negative regulator of FGF and ERK signaling (42). In addition, recent findings showed that DUSP6 expression is stimulated by FGF2, FGF4 and FGF8 via the activation of the ERK pathway (43). Based on these findings, the increased FGF8 expression in several recurrent TD lines may contribute to the high level of DUSP6 expression and the increased levels of DUSP6 may in turn play a role in regulating FGF pathway activity. The absence of DUSP6 overexpression in recurrent subclones Rec3a and Rec3b, which do not overexpress FGF8, provides additional evidence to support this hypothesis.
Currently, even a highly specific targeted therapy, when applied as a monotherapy, is generally not sufficient to eradicate all tumor cells and often leads to tumor recurrence. Based on genomic profiling of resistant and recurrent tumors, combined therapies have been developed for a number of cancer types to reduce the occurrence of resistance and recurrence (44,45). For instance, recent studies have shown that a MET inhibitor combined with a third generation EGFR inhibitor impairs the growth of cancer cells harboring MET amplification (46). Our in vitro data indicate that FGF8 is overexpressed in human FP RMS lines and that FGF8 contributes to the proliferation and tumorigenicity of FP RMS tumors. In addition, FGF8 is able to transform RMS cells in the absence of P3F expression, thus simulating a recurrence mechanism after a P3F-targeted therapy. It is important to realize that, although we can simulate therapy targeting P3F in our myoblast model, the current therapies used to treat FP RMS do not specifically target the P3F fusion. A number of targeted agents have been shown to have activity targeting the P3F fusion (47,48) and thus we anticipate that anti-P3F therapy will be realized in the future. Based on these combined findings, FGF8 may constitute a potential therapeutic target for which a targeted therapy can be combined with a P3F targeted therapy in FP RMS. A growth factor-targeted therapy has been reported recently in a phase 1b trial of non-small cell lung cancer resistant to anti-EGFR treatment and displaying high levels of hepatocyte growth factor (HGF) (49); in this trial, the combination of anti-HGF monoclonal antibody (Ficlatuzumab) with an EGFR inhibitor has shown promising results (50). The possibility of targeting FGF8 was demonstrated in prostate cancer, where FGF8 function was successfully neutralized in vitro and in vivo using monoclonal antibodies (49,51). We therefore propose that the development of anti-P3F therapy delivered along with FGF-signaling inhibitors may be of substantial benefit and avoid relapses in FP RMS cancer. In this way, the findings from this study may ultimately help to effectively treat FP RMS cancers by accelerating the development of new strategies to target predicted resistance mechanisms following application of therapies directed at the P3F fusion.
Supplementary Material
Statement of significance.
In a model of cancer initiated by a fusion transcription factor, constitutive activation of a downstream transcriptional target leads to fusion oncoprotein-independent recurrences, thereby highlighting a novel progression mechanism and therapeutic target.
Acknowledgments
F.G. Barr received support from the intramural program of the National Cancer Institute. This work was supported in part by the Joanna McAfee Childhood Cancer Foundation. S. Boudjadi received a postdoctoral fellowship from the Fonds de recherche du Québec – Santé (FRQS).
Footnotes
Conflict of interest statement
The authors declare no potential conflicts of interest.
References
- 1.Ognjanovic S, Linabery AM, Charbonneau B, Ross JA. Trends in childhood rhabdomyosarcoma incidence and survival in the United States, 1975–2005. Cancer 2009;115:4218–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham J, Nunez-Alvarez Y, Hettmer S, Carrio E, Chen HI, Nishijo K, et al. Lineage of origin in rhabdomyosarcoma informs pharmacological response. Genes Dev 2014;28:1578–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr FG, Galili N, Holick J, Biegel JA, Rovera G, Emanuel BS. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat Genet 1993;3:113–7 [DOI] [PubMed] [Google Scholar]
- 4.Barr FG. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene 2001;20:5736–46 [DOI] [PubMed] [Google Scholar]
- 5.Xia SJ, Holder DD, Pawel BR, Zhang C, Barr FG. High expression of the PAX3-FKHR oncoprotein is required to promote tumorigenesis of human myoblasts. Am J Pathol 2009;175:2600–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandey PR, Chatterjee B, Olanich ME, Khan J, Miettinen MM, Hewitt SM, et al. PAX3-FOXO1 is essential for tumour initiation and maintenance but not recurrence in a human myoblast model of rhabdomyosarcoma. J Pathol 2017;241:626–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olanich ME, Sun W, Hewitt SM, Abdullaev Z, Pack SD, Barr FG. CDK4 Amplification Reduces Sensitivity to CDK4/6 Inhibition in Fusion-Positive Rhabdomyosarcoma. Clin Cancer Res 2015;21:4947–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–8 [DOI] [PubMed] [Google Scholar]
- 9.Boudjadi S, Carrier JC, Groulx JF, Beaulieu JF. Integrin alpha1beta1 expression is controlled by c-MYC in colorectal cancer cells. Oncogene 2016;35:1671–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gryder BE, Yohe ME, Chou HC, Zhang X, Marques J, Wachtel M, et al. PAX3-FOXO1 Establishes Myogenic Super Enhancers and Confers BET Bromodomain Vulnerability. Cancer Discov 2017;7:884–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pei Y, Sun X, Guo X, Yin H, Wang L, Tian F, et al. FGF8 promotes cell proliferation and resistance to EGFR inhibitors via upregulation of EGFR in human hepatocellular carcinoma cells. Oncol Rep 2017;38:2205–10 [DOI] [PubMed] [Google Scholar]
- 13.Liu R, Huang S, Lei Y, Zhang T, Wang K, Liu B, et al. FGF8 promotes colorectal cancer growth and metastasis by activating YAP1. Oncotarget 2015;6:935–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka A, Kamiakito T, Takayashiki N, Sakurai S, Saito K. Fibroblast growth factor 8 expression in breast carcinoma: associations with androgen receptor and prostate-specific antigen expressions. Virchows Arch 2002;441:380–4 [DOI] [PubMed] [Google Scholar]
- 15.Daphna-Iken D, Shankar DB, Lawshe A, Ornitz DM, Shackleford GM, MacArthur CA. MMTV-Fgf8 transgenic mice develop mammary and salivary gland neoplasia and ovarian stromal hyperplasia. Oncogene 1998;17:2711–7 [DOI] [PubMed] [Google Scholar]
- 16.Lui VW, Yau DM, Cheung CS, Wong SC, Chan AK, Zhou Q, et al. FGF8b oncogene mediates proliferation and invasion of Epstein-Barr virus-associated nasopharyngeal carcinoma cells: implication for viral-mediated FGF8b upregulation. Oncogene 2011;30:1518–30 [DOI] [PubMed] [Google Scholar]
- 17.Beltran H, Tomlins S, Aparicio A, Arora V, Rickman D, Ayala G, et al. Aggressive variants of castration-resistant prostate cancer. Clin Cancer Res 2014;20:2846–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doctor SM, Tsao CK, Godbold JH, Galsky MD, Oh WK. Is prostate cancer changing?: evolving patterns of metastatic castration-resistant prostate cancer. Cancer 2014;120:833–9 [DOI] [PubMed] [Google Scholar]
- 19.Mitra D, Brumlik MJ, Okamgba SU, Zhu Y, Duplessis TT, Parvani JG, et al. An oncogenic isoform of HER2 associated with locally disseminated breast cancer and trastuzumab resistance. Mol Cancer Ther 2009;8:2152–62 [DOI] [PubMed] [Google Scholar]
- 20.Rexer BN, Arteaga CL. Intrinsic and acquired resistance to HER2-targeted therapies in HER2 gene-amplified breast cancer: mechanisms and clinical implications. Crit Rev Oncog 2012;17:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi YH, Yu AM. ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr Pharm Des 2014;20:793–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apperley JF. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol 2007;8:1018–29 [DOI] [PubMed] [Google Scholar]
- 23.Jang JW, Boxer RB, Chodosh LA. Isoform-specific ras activation and oncogene dependence during MYC- and Wnt-induced mammary tumorigenesis. Mol Cell Biol 2006;26:8109–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med 2001;7:235–9 [DOI] [PubMed] [Google Scholar]
- 25.Hancock BA, Chen YH, Solzak JP, Ahmad MN, Wedge DC, Brinza D, et al. Profiling molecular regulators of recurrence in chemorefractory triple-negative breast cancers. Breast Cancer Res 2019;21:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schafer N, Gielen GH, Rauschenbach L, Kebir S, Till A, Reinartz R, et al. Longitudinal heterogeneity in glioblastoma: moving targets in recurrent versus primary tumors. J Transl Med 2019;17:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Kessler P, Yeger H, Alami J, Reeve AE, Heathcott R, et al. A gene expression signature for relapse of primary wilms tumors. Cancer Res 2005;65:2592–601 [DOI] [PubMed] [Google Scholar]
- 28.Kwon SM, Kang SH, Park CK, Jung S, Park ES, Lee JS, et al. Recurrent Glioblastomas Reveal Molecular Subtypes Associated with Mechanistic Implications of Drug-Resistance. PLoS One 2015;10:e0140528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh N, Ohta H, Nakayama Y, Konishi M. Roles of FGF Signals in Heart Development, Health, and Disease. Front Cell Dev Biol 2016;4:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010;10:116–29 [DOI] [PubMed] [Google Scholar]
- 31.Kawakami Y, Rodriguez-Leon J, Koth CM, Buscher D, Itoh T, Raya A, et al. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat Cell Biol 2003;5:513–9 [DOI] [PubMed] [Google Scholar]
- 32.Mattila MM, Harkonen PL. Role of fibroblast growth factor 8 in growth and progression of hormonal cancer. Cytokine Growth Factor Rev 2007;18:257–66 [DOI] [PubMed] [Google Scholar]
- 33.Tanaka A, Furuya A, Yamasaki M, Hanai N, Kuriki K, Kamiakito T, et al. High frequency of fibroblast growth factor (FGF) 8 expression in clinical prostate cancers and breast tissues, immunohistochemically demonstrated by a newly established neutralizing monoclonal antibody against FGF 8. Cancer Res 1998;58:2053–6 [PubMed] [Google Scholar]
- 34.Sato T, Oshima T, Yoshihara K, Yamamoto N, Yamada R, Nagano Y, et al. Overexpression of the fibroblast growth factor receptor-1 gene correlates with liver metastasis in colorectal cancer. Oncol Rep 2009;21:211–6 [PubMed] [Google Scholar]
- 35.Harpain F, Ahmed MA, Hudec X, Timelthaler G, Jomrich G, Mullauer L, et al. FGF8 induces therapy resistance in neoadjuvantly radiated rectal cancer. J Cancer Res Clin Oncol 2019;145:77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erdem ZN, Schwarz S, Drev D, Heinzle C, Reti A, Heffeter P, et al. Irinotecan Upregulates Fibroblast Growth Factor Receptor 3 Expression in Colorectal Cancer Cells, Which Mitigates Irinotecan-Induced Apoptosis. Transl Oncol 2017;10:332–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 2005;8:197–209 [DOI] [PubMed] [Google Scholar]
- 38.Tanaka A, Miyamoto K, Minamino N, Takeda M, Sato B, Matsuo H, et al. Cloning and characterization of an androgen-induced growth factor essential for the androgen-dependent growth of mouse mammary carcinoma cells. Proc Natl Acad Sci U S A 1992;89:8928–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gnanapragasam VJ, Robson CN, Neal DE, Leung HY. Regulation of FGF8 expression by the androgen receptor in human prostate cancer. Oncogene 2002;21:5069–80 [DOI] [PubMed] [Google Scholar]
- 40.Liu F, You X, Wang Y, Liu Q, Liu Y, Zhang S, et al. The oncoprotein HBXIP enhances angiogenesis and growth of breast cancer through modulating FGF8 and VEGF. Carcinogenesis 2014;35:1144–53 [DOI] [PubMed] [Google Scholar]
- 41.Hanna JA, Garcia MR, Lardennois A, Leavey PJ, Maglic D, Fagnan A, et al. PAX3-FOXO1 drives miR-486–5p and represses miR-221 contributing to pathogenesis of alveolar rhabdomyosarcoma. Oncogene 2018;37:1991–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li C, Scott DA, Hatch E, Tian X, Mansour SL. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development 2007;134:167–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmad MK, Abdollah NA, Shafie NH, Yusof NM, Razak SRA. Dual-specificity phosphatase 6 (DUSP6): a review of its molecular characteristics and clinical relevance in cancer. Cancer Biol Med 2018;15:14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oxnard GR, Cantarini M, Frewer P, Hawkins G, Peters J, Howarth P, et al. SAVANNAH: A Phase II trial of osimertinib plus savolitinib for patients (pts) with EGFR-mutant, MET-driven (MET+), locally advanced or metastatic non-small cell lung cancer (NSCLC), following disease progression on osimertinib. Journal of Clinical Oncology 2019;37:TPS9119–TPS [Google Scholar]
- 45.Yu C, Liu X, Yang J, Zhang M, Jin H, Ma X, et al. Combination of Immunotherapy With Targeted Therapy: Theory and Practice in Metastatic Melanoma. Front Immunol 2019;10:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, Yang S, Wang K, Sun SY. MET inhibitors for targeted therapy of EGFR TKI-resistant lung cancer. J Hematol Oncol 2019;12:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amstutz R, Wachtel M, Troxler H, Kleinert P, Ebauer M, Haneke T, et al. Phosphorylation regulates transcriptional activity of PAX3/FKHR and reveals novel therapeutic possibilities. Cancer Res 2008;68:3767–76 [DOI] [PubMed] [Google Scholar]
- 48.Jothi M, Mal M, Keller C, Mal AK. Small molecule inhibition of PAX3-FOXO1 through AKT activation suppresses malignant phenotypes of alveolar rhabdomyosarcoma. Mol Cancer Ther 2013;12:2663–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yano S, Yamada T, Takeuchi S, Tachibana K, Minami Y, Yatabe Y, et al. Hepatocyte growth factor expression in EGFR mutant lung cancer with intrinsic and acquired resistance to tyrosine kinase inhibitors in a Japanese cohort. J Thorac Oncol 2011;6:2011–7 [DOI] [PubMed] [Google Scholar]
- 50.Tan EH, Lim WT, Ahn MJ, Ng QS, Ahn JS, Shao-Weng Tan D, et al. Phase 1b Trial of Ficlatuzumab, a Humanized Hepatocyte Growth Factor Inhibitory Monoclonal Antibody, in Combination With Gefitinib in Asian Patients With NSCLC. Clin Pharmacol Drug Dev 2018;7:532–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimada N, Ishii T, Imada T, Takaba K, Sasaki Y, Maruyama-Takahashi K, et al. A neutralizing anti-fibroblast growth factor 8 monoclonal antibody shows potent antitumor activity against androgen-dependent mouse mammary tumors in vivo. Clin Cancer Res 2005;11:3897–904 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.