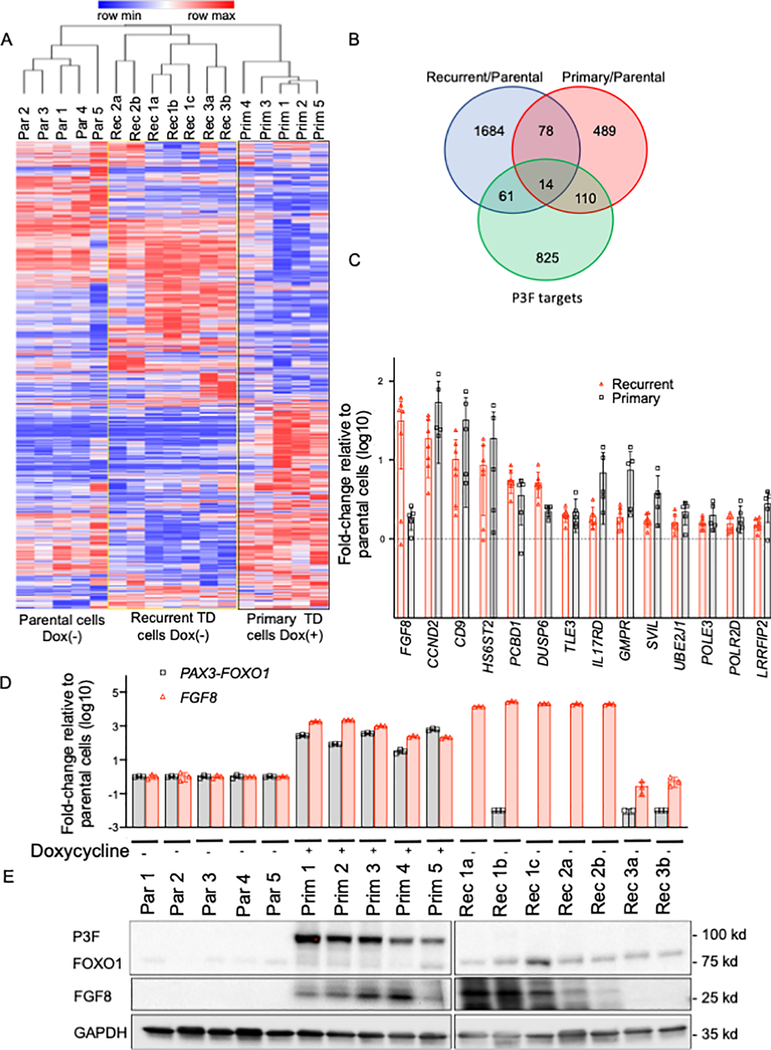
Figure 1. Differences in expression between recurrent and primary TD cells.
A. Hierarchical clustering shows differentially expressed genes between unstimulated parental (Par) cells and recurrent (Rec) TD cells and doxycycline-treated primary (Prim) TD cells. Each column represents an individual cell line (indicated by number) and/or subclone (indicated by letter). The blue and red lines in the heat map indicate low and high gene expression levels according to the scale shown at the top of the heat map. B. Venn diagram showing overlap of the upregulated genes in recurrent TD cells, doxycycline-treated primary TD cells and P3F target genes (11). Differentially upregulated genes were selected using a false discovery adjusted p-value of 0.01 and a 1.5-fold change cut off. C. Analysis of the 14 P3F target genes (described in B) that are significantly upregulated in both recurrent and primary TD cells compared to parental cells. For both primary and recurrent TD cells, array data was used to calculate the expression increase relative to parental cells. Genes are ranked based on the expression increase in recurrent TD cells. P3F (gray bars) and FGF8 (red bars). D. P3F and FGF8 mRNA expression levels in parental cells, primary TD cells and recurrent TD cells. Some cells were treated with doxycycline, as shown. P3F and FGF8 mRNA expression was assayed by real-time RT-PCR and normalized for GAPDH expression by the ΔΔCt method. The data are expressed as the mean +/− SE of 3 replicates. E. P3F and FGF8 protein expression in the three groups of cells (as described in part D). Western blot analysis was performed using GAPDH expression as a control for protein loading.