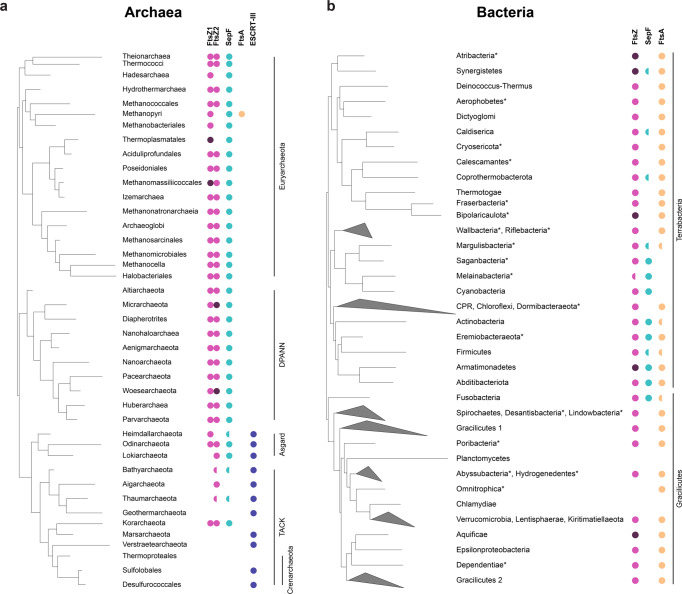
Fig. 5. SepF is widely present in Archaea and co-occurs with FtsZ.
a Distribution of FtsZ1, FtsZ2, SepF, FtsA and ESCRT-III (CdvB) homologs on a schematic reference phylogeny of the Archaea. FtsZ1 and FtsZ2 homologs are present in most archaeal lineages (magenta), and frequently in two or more copies each (dark magenta). Semicircles indicate that the corresponding protein could not be identified in all the taxa of the corresponding clade and may be either due to true absences or partial genomes. The presence of SepF (turquoise) correlates to that of FtsZ in the majority of taxa. A FtsA homolog was only identified in Methanopyri (orange). Homologs of ESCRT-III (CdvB and homologs) (purple) are only present in the Asgard superphylum and in most representatives of the TACK superphylum, except for Korarchaeota and Thermoproteales. For full data see Supplementary Data 1. b Distribution of FtsZ, SepF and FtsA homologs on a schematic reference phylogeny of Bacteria. FtsZ (magenta) is present in most bacterial phyla, with the exception of some phyla within the PVC superphylum (Planctomycetes, Omnitrophica and Chlamydiae), which are known to have specific FtsZ-less cytokinesis. Both SepF and FtsA are also absent in Planctomycetes and Chlamydiae. Two copies of FtsZ can be identified in Atribacteria, Synergistetes, Biopolaricaulota, Armatimonadetes and Aquificae (dark magenta). SepF is present in some phyla (Synergistetes, Caldiserica, Coprothermobacterota, Margulisbacteria, Melainabacteria, Cyanobacteria, Actinobacteria, Eremiobacteraeota, Firmicutes, Armatimonadetes, Abditibacteriota and Fusobacteria), most belonging to the Terrabacteria. In contrast, all Gracilicutes except Fusobacteria, and the remaining Terrabacteria phyla have only FtsA, while Cyanobacteria, Melainabacteria and Saganbacteria have only SepF. Finally, some members of Synergistetes, Caldiserica, Coprothermobacterota, Margulisbacteria, Actinobacteria, Eremiobacteraeota, Firmicutes, Armatimonadetes, Abditibacteriota and Fusobacteria have both FtsA and SepF. Semicircles indicate that the corresponding protein could not be identified in all the analyzed taxa in the linage displayed. * indicates uncultured Candidate phyla for which many genomes are incomplete. These phyla were not included in phylogenetic reconstructions. For full data see Supplementary Data 2.