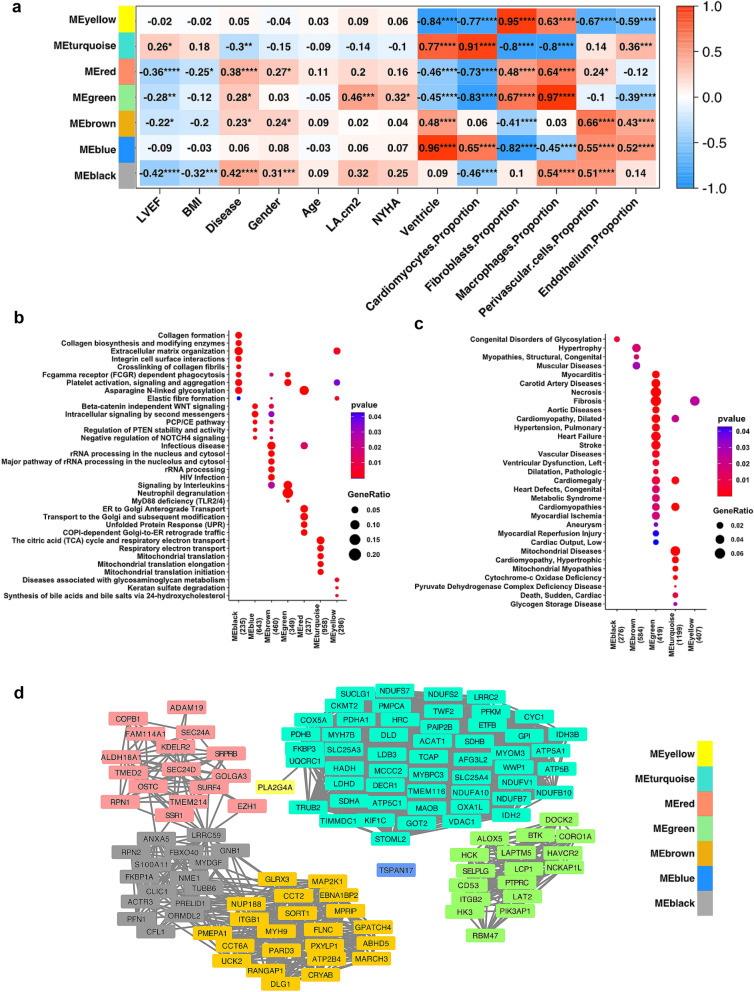
Figure 7.
Network analysis revealed key modules and genes altered in HFrEF-diseased heart. (a) WGCNA analysis classified genes into 7 major modules. The heatmap shows the correlation score between each of the gene modules with the analyzed sample and clinical feature groups (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001). Comparison of over-representative (b) Reactome pathways and (c) cardiometabolic diseases associated MeSH terms among modules determined by WGCNA analysis. The size of the dot reflects the GeneRatio (the proportion of genes from list of interests that maps to the pathway) and the color of the dot indicates increasing significance of the enriched pathways from blue to red. Number of chamber enriched genes that overlap with the over-representative pathways are shown in parenthesis. Significantly enriched terms are presented in (b,c) and significance is indicated as p < 0.05. (d) Disease-associated hub genes (p < 0.05) with top connectivity are visualized by Cytoscape. Color of each node corresponds with the module color in (a).