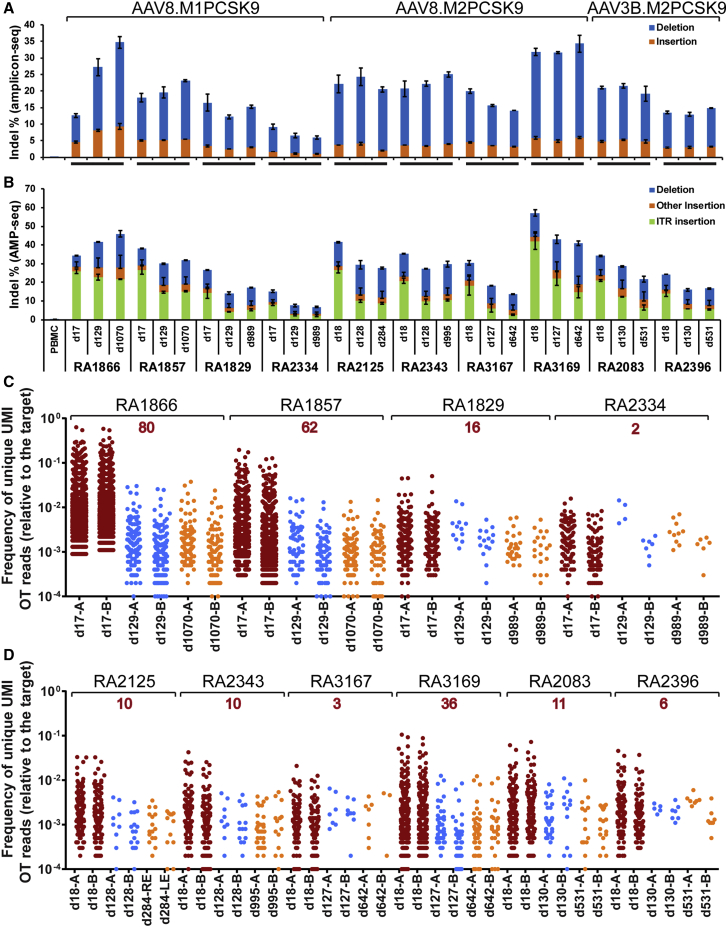
Figure 2.
Analyses of on-target and OT editing in consecutive macaque liver biopsy and necropsy samples collected at various time points after vector administration
Each biopsy sample was evaluated by two independent genomic DNA isolations from the biopsy sample followed by next-generation sequencing (NGS). (A and B) On-target editing analyzed by amplicon-seq (A) or AMP-seq (B). Indels in the liver of RA2125 collected at necropsy were analyzed in eight samples, two each from the left, right, middle, and caudate lobes. The means ± SEM are shown. (C and D) Off-target (OT) editing evaluated by ITR-seq on AAV8.M1PCSK9-treated liver biopsy samples (C) or AAV.M2PCSK9-treated liver samples (D). Each dot represents a unique potential OT site detected by ITR-seq and its relative frequency with respect to on-target editing. The number in red above the animal ID indicates the number of OT sites detected in samples from all three time points of the same animal. Detailed OT information can be found in Data S1. UMI, unique molecular identifier.