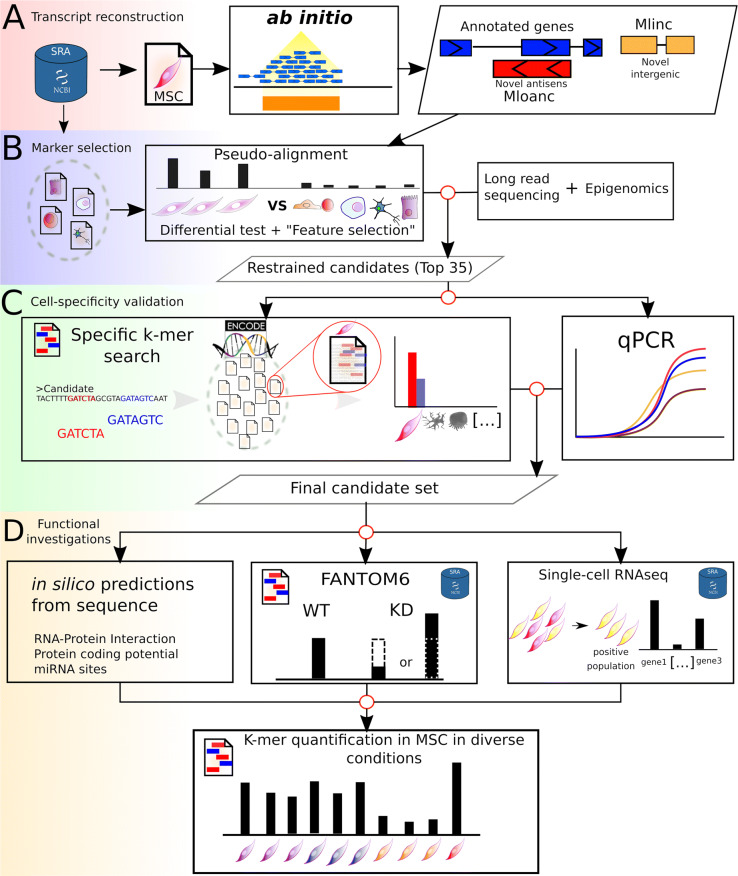
Fig. 1.
Flowchart representation of the pipeline used in this study. The 4 steps of the flowchart are described. a Ab initio reconstruction of transcript expressed in MSCs from SRA dataset and creation of a reference (GTF+fasta) for quantification of Ensembl annotated genes, unannotated intergenic (Mlincs) and unannotated overlapping antisens (Mloanc). The results are shown in Fig. 2. b Differential Analysis for the selection of MSC markers (restrained candidates set) with Kallisto pseudoalignement and Sleuth differential test followed by feature selection by random forest with Boruta package. Long-read sequencing and active transcription in MSCs by epigenetic marks information completed the selection step (see Figs. 2 and 3). c Validation of cell expression specificity of the candidates by k-mer quantification in ENCODE RNAseq datasets (see Additional file 8 for the list of data) and qPCR validation. The results are presented in Fig. 4. d Functional investigations were performed with in silico prediction methods from the sequence of candidates, followed by k-mer quantification with FANTOM6 dataset, single-cell RNAseq and selected MSC conditions. K-mer quantification phases are shown by corresponding icons (Figs. 5 and 6)