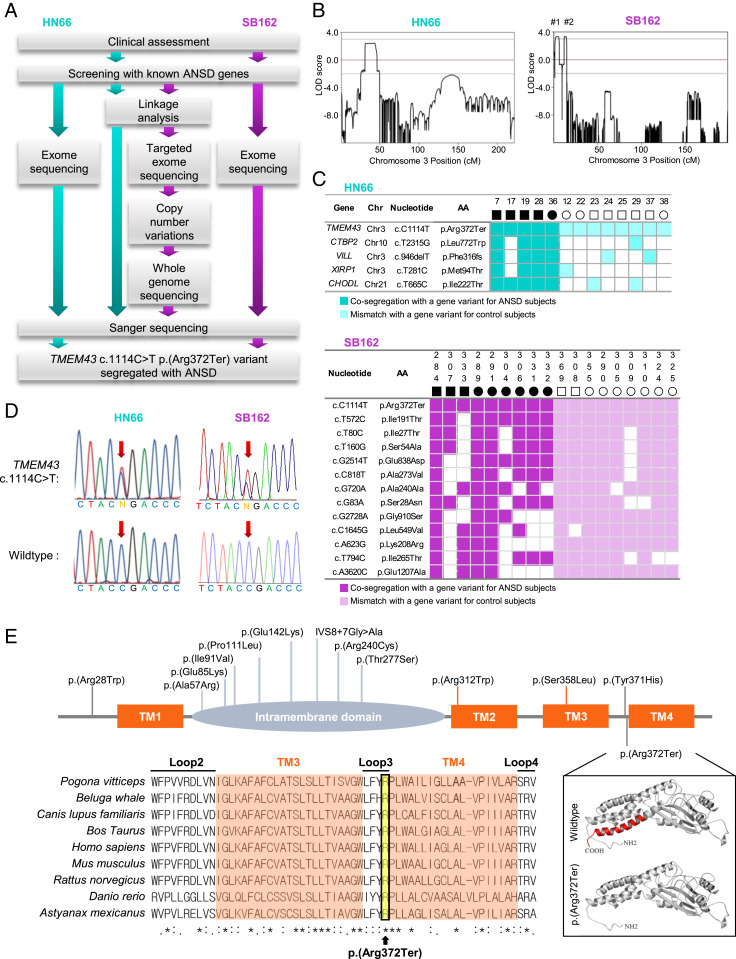
Fig. 2.
A p.(Arg372Ter) variant of TMEM43 is the causative variant of ANSD in family HN66 and SB162. (A) Overall workflow of variant detection in HN66 and SB162. (B) Merlin multipoint linkage analysis demonstrates LOD scores greater than 3.0 on chromosome 3p25.1. Region #1 (Chr. 3: 1,946,000 to 5,956,000), region #2 (Chr. 3:11,883,000 to 14,502,000). (C) Genetic candidates of the ANSD after exome sequencing of genomic DNA from three subjects (#22, #28, and #36) in HN66 and four subjects (#289, #290, #291, and #284) in SB162. Subsequent segregation study with additional subjects to identify TMEM43 variant in HN66 and SB162 with full-match was performed. Cosegregation with a gene variant for ANSD subjects in HN66 and SB162 are indicated as cyan and purple box, respectively. Mismatch with a gene variant for control subjects are lightly colored. Chr; chromosome, AA; amino acid. (D) Representative DNA sequence chromatograms of c.1114C > T variant in TMEM43. Red arrow signifies allele c.1114C. (E) The variant spectrum of TMEM43 associated with ARVC and Emery–Dreifuss muscular dystrophy 7 (EDMD7) reported in human gene mutation database (HGMD) (Top) and the variants identified in the present study [Bottom, p.(Arg372Ter)]. The conservative amino acid sequences of TMEM43 are shown below. Asterisks, identical amino acids; orange box, TM domains; yellow box, p.(Arg372Ter). A protein structure modeling of WT human TMEM43 and the variant TMEM43 lacking TM4 (red) is shown. TM, transmembrane domain.