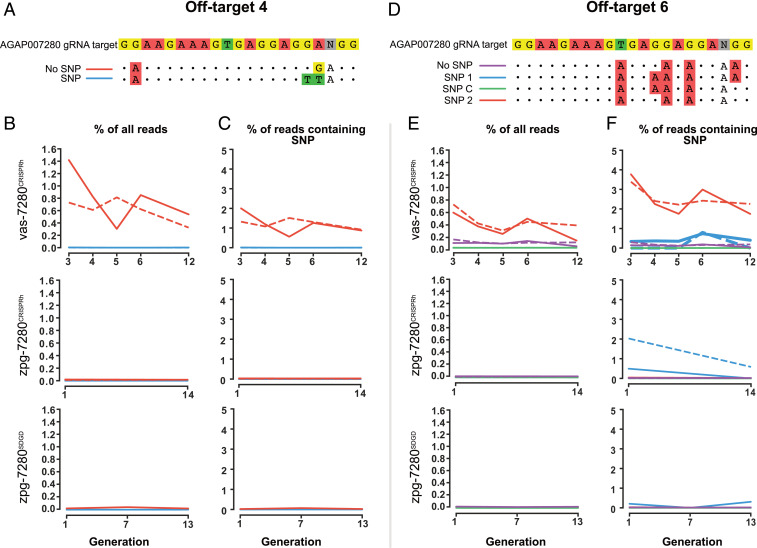
Fig. 3.
The impact of genetic variation upon CRISPR-Cas9 off-target cleavage. (A) Off-target 4 contained two alleles. (B) Indels were detected in one allele (No SNP) within vas-7280CRISPRh population, with no significant indels detected for zpg-7280CRISPRh or zpg-7280SDGD. This represents editing as a percentage of all reads that align to the off-target site (C) Levels of indels detected are displayed as a percentage of reads that align to each allele, respectively. (D) Off-target 6 contained four alleles. (E) Indels were detected in one allele (SNP 2) which contained an NGG PAM. (F) SNP 2 showed indels for vas2-CRISPRh. Indels found for SNP 1 in zpg-7280CRISPRh are not evidence of genuine off-targeting editing, but artifacts of very low number of reads aligning to this allele. Cage 1 (solid line) and cage 2 (dash line), with zpg-7280SDGD containing one biological replicate only (due to the other cage’s population crashing at G8, as intended as part of the study).