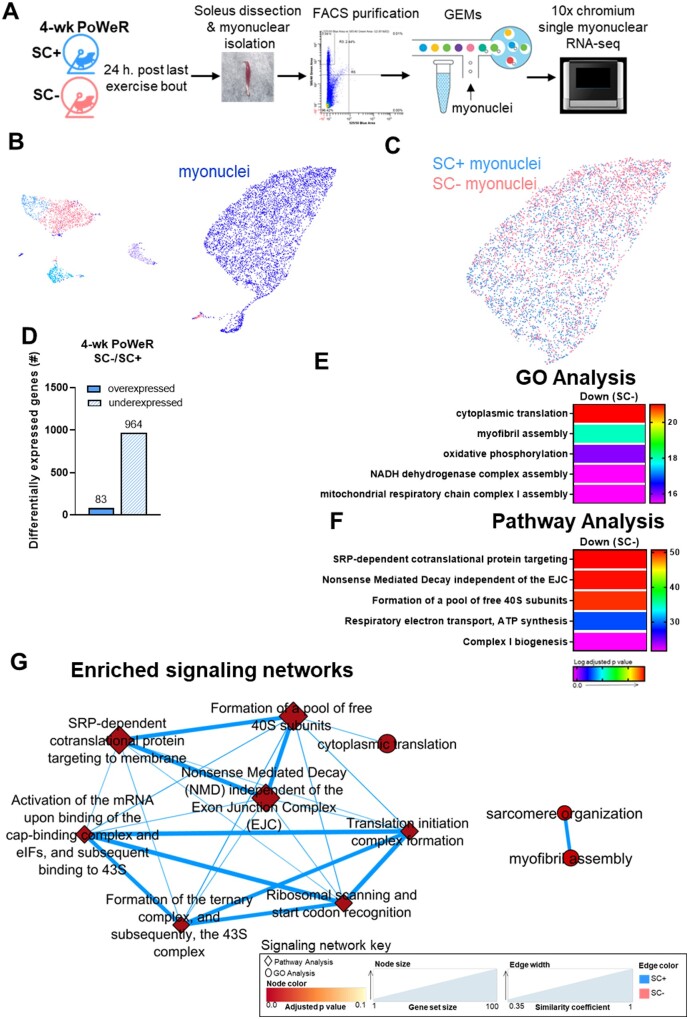
Figure 6.
Myonuclear transcriptome reveals a greater downregulation of oxidative, ribosomal, and sarcomeric pathways in the absence of satellite cells in the soleus. (A) Schematic of myonuclear isolation and sequencing from skeletal muscle following PoWeR. (B) Unbiased cluster of smnRNA-seq data, including all cell types and myonuclei from SC+ and SC− mice represented on a UMAP plot. (C) Distinct myonuclear cluster showing myonuclei from SC+ muscle in blue and SC− muscle in pink. (D) Bar graph showing the number of over and underexpressed genes in the soleus after 4-weeks of PoWeR in SC+ skeletal muscle when compared to SC− skeletal muscle. The most highly enriched (E) biological processes (GO Analysis) and (F) Reactome pathways (Pathway Analysis). (G) Translation and sarcomeric signaling networks.