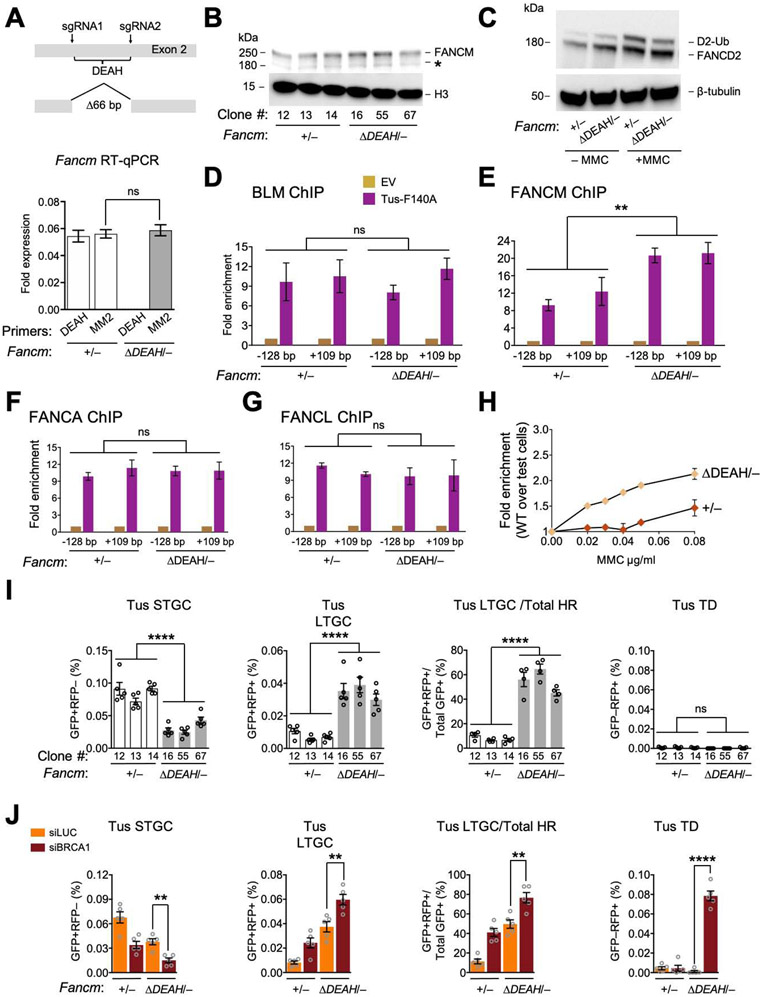
Figure 5. FANCM ATP hydrolysis mutants are defective for FANCM-mediated stalled fork repair.
See also Figures S5 and S6. A. FancmΔDEAH allele and RT qPCR analysis of MM2 and DEAH encoding mRNA in Fancm+/− and FancmΔDEAH/− clones. Data shows mean ± SD. Analysis by Student’s t-test (n=3). B. Immunoblot of chromatin-extracted FANCM in Fancm+/− and FancmΔDEAH/− clones. C. Immunoblot showing FANCD2 ubiquitination in Fancm+/− and FancmΔDEAH clones. D-G. ChIP analysis of BLM (D), FANCM (E), FANCA (F) and FANCL (G) at Tus/Ter in Fancm+/− and FancmΔDEAH/− cells. Data shows mean ± SD. Analysis by one-way ANOVA (n=3). H. Proliferative competition assay in presence of MMC, measuring enrichment of GFP+ Fancm+/− vs. GFP− FancmΔDEAH/−cells (n=3). Error bars: SD. I. Tus/Ter-induced HR in Fancm+/− (white) clones vs. FancmΔDEAH/− (gray) clones. Data shows mean ± SEM. Analysis by one-way ANOVA (n=5). J. Tus/Ter-induced repair in Fancm+/− vs. FancmΔDEAH/− clones co-transfected with Tus and siRNAs as shown. Data shows mean ± SEM. Analysis by Student’s t-test (n=5).