FIGURE 1.
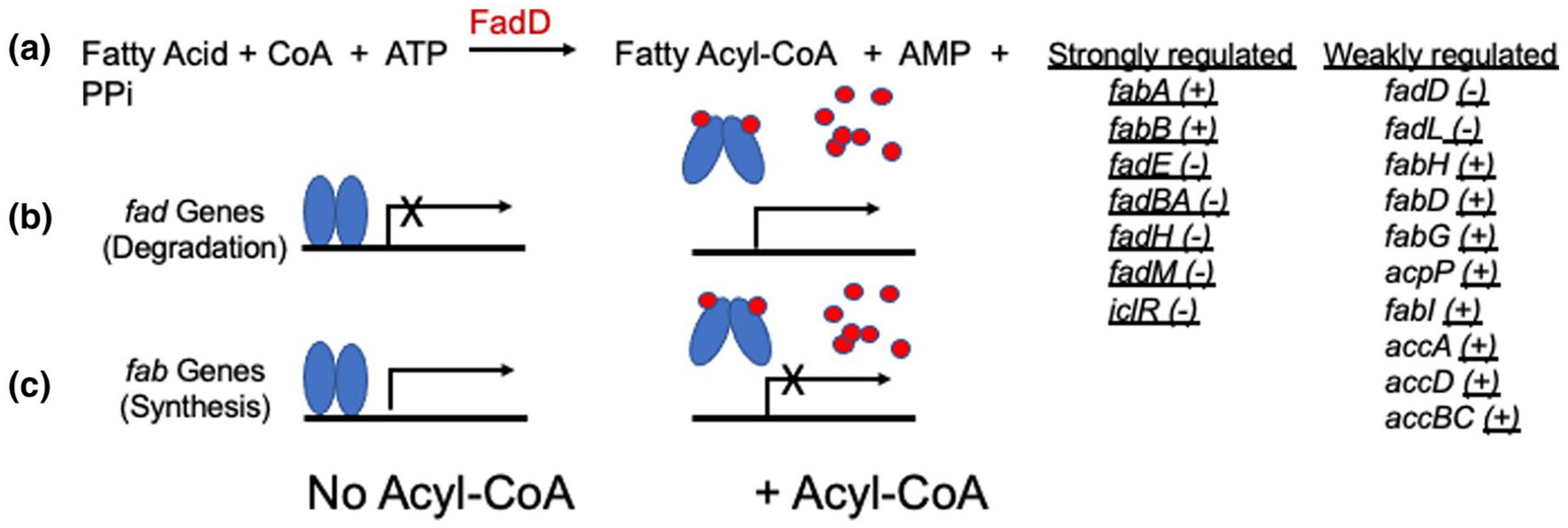
Fad R Regulates Two Opposing Pathways in E. coli. Fad R is a repressor of the fad genes and an activator of the fab genes. When present, environmental long-chain fatty acids are converted to their acyl-CoA thioesters after entering the cytosol. The acyl-CoAs (red dots) bind to FadR (blue ovals) resulting in the formation of a helix piston that spreads the N-terminal FadR DNA binding domains thereby eliminating the ability to cooperatively bind to the operator sites of the genes. In the absence of exogenous fatty acids (hence no acyl-CoAs) FadR is bound to the operators of both then fad and fab genes. The strongly regulated genes (+ and − denote positive and negative regulation, respectively) are those regulated by wild-type levels of FadR whereas the weakly related genes fall into two classes. The fatty acid uptake genes (fadD and fadL) are weakly negatively regulated by wild-type FadR whereas the fab, acc, and acpP genes show positive regulation only when FadR is overexpressed. Note that the genes of the anaerobic fatty acid degradation pathway are not under FadR regulation (Campbell et al., 2003)
