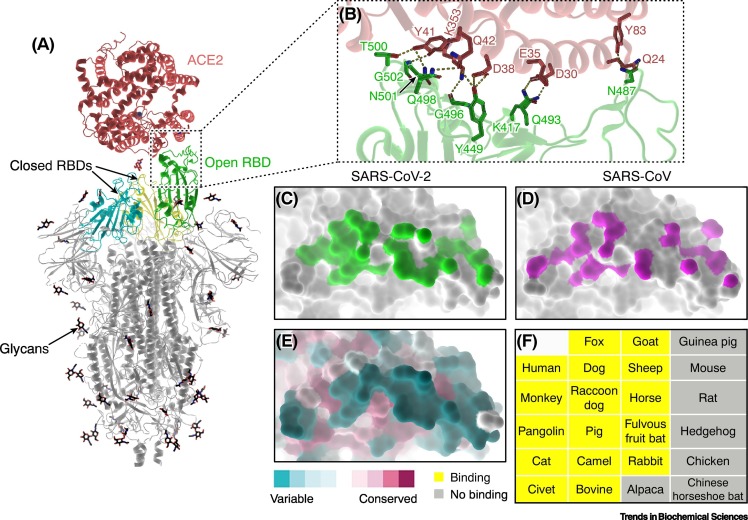
Figure 3.
Interactions between SARS-CoV-2 spike (S) protein and ACE2.
(A) Overall structure of a trimeric S bound to ACE2 (PDB: 7KNB). ACE2 binds to the RBD of S in the open conformation, while the other two closed RBDs are inaccessible by the receptor. (B) Close-up view of the S–ACE2 contacting interface (PDB: 6LZG). The key interacting residues are shown as sticks. Hydrogen bonds and salt bridges are represented by dashed lines. (C,D) The S-binding footprint on ACE2 for SARS-CoV-2 (C; PDB: 6LZG) and SARS-CoV (D; PDB: 2AJF). The interaction between SARS-CoV-2 (green) and ACE2 involves more atomic contacts than for SARS-CoV (magenta) [19., 20., 21.,28]. (E) Conservation of the S-binding interface on ACE2. The interface for SARS-CoV-2 S binding displays a high degree of variation among ACE2 orthologs from different animal species. (F) A list of animals tested whose ACE2 can (yellow) or cannot (grey) bind to the RBD of SARS-CoV-2 S protein [29,31,32]. Abbreviations: ACE2, angiotensin-converting enzyme 2; RBD, receptor-binding domain; SARS-CoV-2, severe acute respiratory syndrome virus 2.