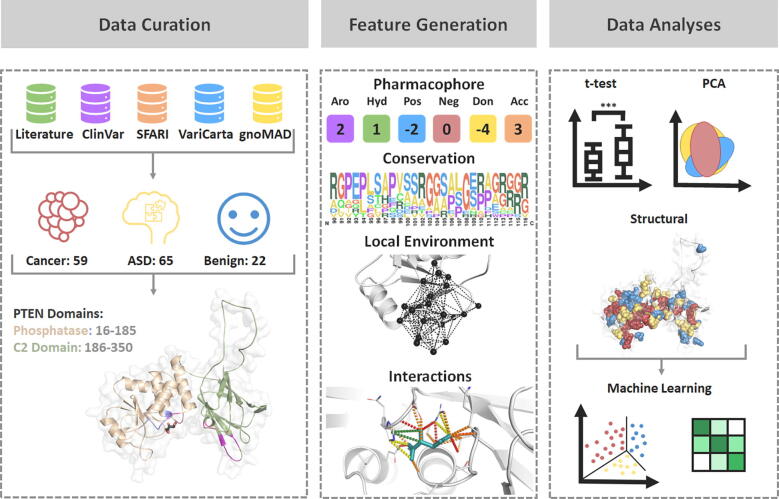
Fig. 2.
Methodology Pipeline followed in this study. Initial mutation curation was carried out to obtain Cancer (n = 59), ASD (n = 65) and Non-Pathogenic (n = 22; labelled as Benign in figure) mutations from five different sources. Data curation also involved processing of the experimental crystal structure to fill in missing residues and model the missing loop (286–309). In silico biophysical tools were then used to measure the effects of mutations on protein structure and function (Feature Generation), which was followed by different structural and statistical analyses and the development of a three-class prediction model.