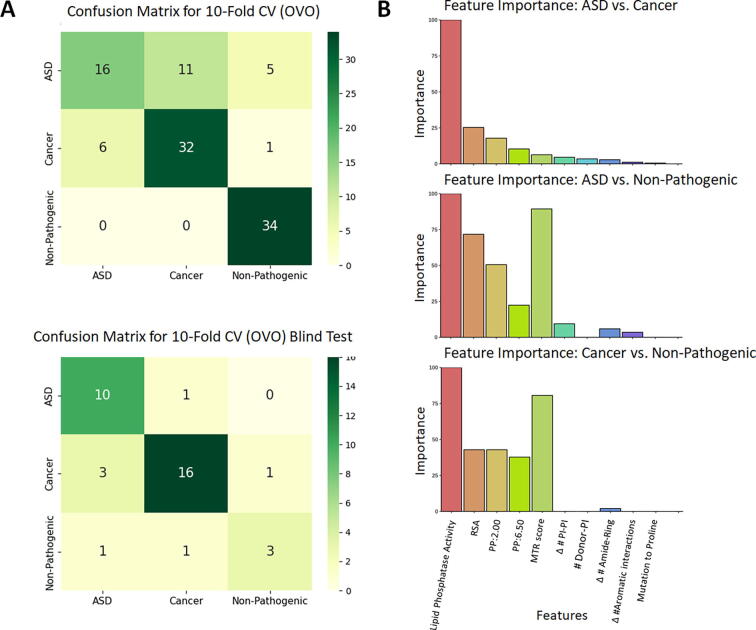
Fig. 5.
Metrics for the chosen model. Model was obtained after greedy feature selection, where the confusion matrices (A) calculated the three-class classifier using the OneVsOne method on the Adaboost algorithm, validated through 10-fold cross validation, with prediction performance of up to 0.68 MCC. Confusion matrices show correctly predicted data points per class, across the diagonal. (B) Observing the contribution of each feature within the estimators of our final model shows that MTR score and RSA are important in identification of disease variants, while changes in lipid phosphatase activity plays an important role in distinguishing between disease outcomes.