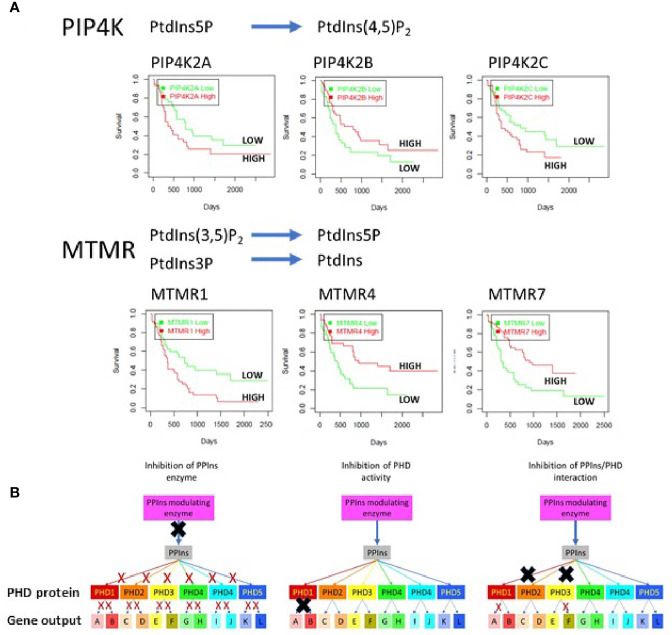
Figure 4.
(A) The TGCA AML patient data base was used to assess survival outcomes with respect to low or high expression of various PPIns modulating enzymes to demonstrate complex signalling and outcomes within enzymes from the same family. As examples we shown that low expression of PIP4K2A, PIP4K2C or MTMR1 is associated with increased survival. Surprisingly however the expression of PIP4K2B, a family member with PIP4K2A and 2C shows survival outcomes that are opposite with respect to its expression. Similar data are observed MTMR4 and 7 where high expression is associated with increased survival in AML. These data suggest that the complexity in PPIns signalling could be exploited to generate modulatory pathways that could be beneficial for AML treatment. (B) A schematic diagram illustrating that PPIns inhibitors could act at 3 points within a PPIns signalling unit: (i) inhibition of the PPIns modulating enzyme will impair all downstream targets and associated gene expression programs (A:L) (ii) inhibition of the activity of a PHD containing protein will likely impair all the genes that the protein regulates (gene A and gene B) or (iii) allosteric inhibition of in the interaction of PPIns with a specific interacting domain will impair selected output which might be tuned to specific pathways and therefore engender less toxic outcomes. In this example inhibition of two PHD domain interactions is shown which impair expression of gene A and gene F.