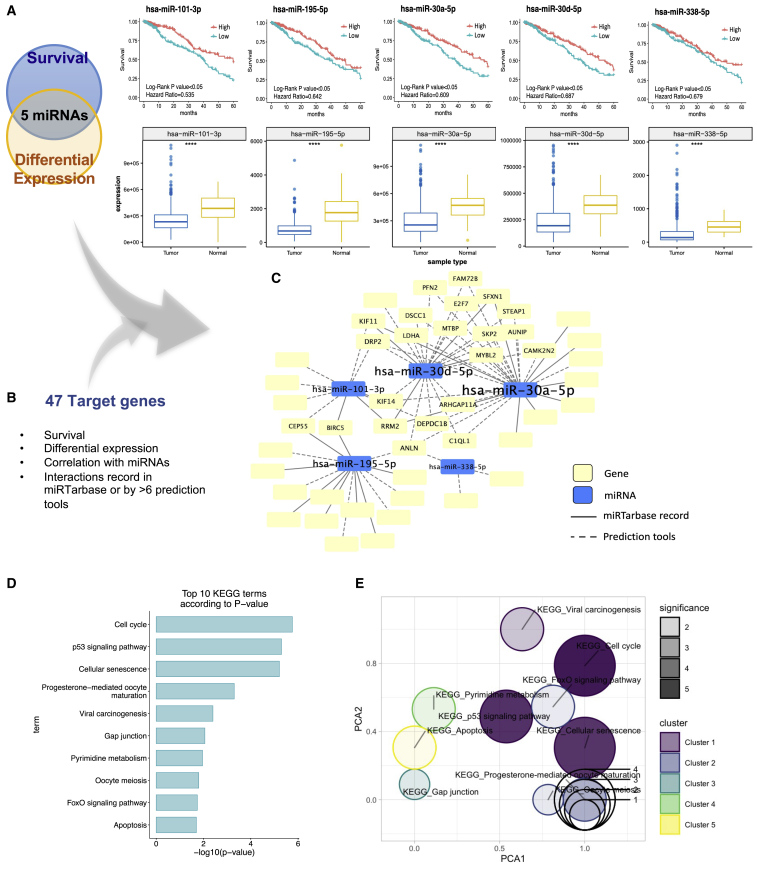
Figure 1.
Defining clinically relevant miRNAs through bioinformatics integrative analysis
(A) Clinically relevant miRNA candidates were filtered out by both significance in terms of survival and differential expression in TCGA LUAD dataset. 5-year survival analysis of five miRNA candidates indicated that they were significant, with log-rank p < 0.05 and HR values < 1. The expression level differences of these five miRNA candidates between LUAD tumor and normal samples in TCGA are displayed. The asterisks (∗∗∗∗) denote statistical significance with p < 0.0001. (B) Target genes were defined by their survival significance levels, differential expression levels, correlations with the miRNA candidates, and interactions as indicated in the miRTarbase or by >6 prediction tools. (C) The network displays the interactions between the miRNA candidates and the target genes recorded in the miRTarbase (solid lines) or defined by the prediction tools (dashed lines). (D) The top 10 significant KEGG pathways of the 47 target genes are shown based on the p values of the functional annotation analysis results. (E) A gene set overrepresentation of significant KEGG pathways displays overlapped genes in the different pathways.