Summary
Hepatocellular carcinoma (HCC) is the most common form of primary liver cancer. Its incidence is rising faster than any other cancer in the United States and it remains one of the leading causes of cancer-related deaths worldwide. While advances in massive parallel sequencing and integration of ‘omics information have transformed the field of oncology, tissue access is often limited in HCC and a single biopsy is poorly representative of the known genetic heterogeneity of tumours. Liquid biopsy has emerged as a promising strategy for analysing circulating tumour components including circulating tumour DNA. Cell-free DNA and tumour DNA are derived from necrotic, apoptotic and living eukaryotic cells. The profiling of genetic and epigenetic alterations in circulating cell-free DNA has potential clinical applications including early disease detection, prediction of treatment response and prognostication in real time. Novel biomarker candidates for disease detection and monitoring are under study. Of these, methylation analyses of circulating tumour DNA have shown promising performance for early HCC detection in at-risk patients. Assessments of assay performance in longitudinal validation cohorts are ongoing. Implementation of liquid biopsy for HCC will likely improve upon the current surveillance strategy. This review summarises the most recent developments on the role and utility of circulating cell-free DNA in the detection and management of HCC.
Keywords: cfDNA, ctDNA, circulating biomarker, liver cancer, hepatocellular carcinoma, cell-free nucleic acids
Abbreviations: AFP, alpha-fetoprotein; cfDNA, circulating cell-free DNA; CNV, copy number variants; ctDNA, circulating tumour DNA; HCC, hepatocellular carcinoma; NGS, next-generation sequencing; SNV, single nucleotide variants; WGS, whole genome sequencing
Key points.
-
•Early detection
-
○Early detection of hepatocellular carcinoma leads to early curative treatment and improves survival.
-
○Several methylation panels assayed from plasma DNA have demonstrated high sensitivity and specificity in detecting early disease in at-risk individuals.
-
○
-
•Potential clinical utility of cell-free DNA
-
○Longitudinal prospective studies are ongoing.
-
○Detect cancer in individuals at high risk.
-
○Measure residual disease following surgery, ablation or transplant with risk stratification for adjuvant therapy.
-
○Enable treatment selection.
-
○Elucidate mechanisms of resistance and disease progression.
-
○
Introduction
Hepatocellular carcinoma (HCC), the most common form of primary liver cancer, is one of the leading causes of cancer-related deaths worldwide.1 Each year, more than 800,000 individuals are diagnosed globally. The incidence of HCC is rising faster than that of any other cancer in the United States,2 driven by HCV-associated cirrhosis and the rising prevalence of non-alcoholic fatty liver disease.3 The annual risk of developing HCC in high risk groups, including chronic carriers of HBV and patients with cirrhosis (of infectious, metabolic or alcoholic aetiology) is 2–4% per year.4 Several studies have demonstrated that HCC surveillance is associated with early detection, receipt of curative treatment, and improved survival.5,6 Consequently, for those patients at high risk, clinical practice guidelines recommend biannual HCC surveillance by ultrasound imaging with or without serum alpha-fetoprotein (AFP) testing.7,8 However, several limitations exist with this approach; the aggregate sensitivity is low at 63%, especially for those with early disease within and outside of Milan criteria for liver transplantation, and surveillance is underutilised.9 A recent meta-analysis involving more than 118,000 patients showed a pooled estimate for surveillance use of 24% (95% CI 18.4–30.1), with variable usage depending on the level of care (subspecialty care or primary care).10 Furthermore, the diagnosis of HCC is made using radiological or histological criteria. Liver biopsy is invasive, poorly reflects the temporal and spatial heterogeneity within the tumour, and is often not available. This presents a large window of opportunity for the development of novel biomarkers that can detect HCC early and can accurately predict outcomes.
Currently, liver resection and local ablation remain the mainstays of curative therapy, as liver transplant is limited by the size of the donor pool and stringent eligibility criteria.11,12 After resection, recurrence within the residual cirrhotic liver is high, with more than 50% of patients developing HCC recurrence within 3 years and 70–80% recurring within 5 years; the 5-year survival rate for these patients is approximately 40–50%.[13], [14], [15], [16], [17] Over the last few years, significant advances have been made in the management of patients with advanced HCC. Since 2007, sorafenib18 – an oral multikinase inhibitor – and subsequently lenvatinib,19 have been the first-line systemic therapies for patients with advanced HCC. Recently, atezolizumab – an anti-PD-L1 antibody – in combination with bevacizumab – a monoclonal anti-VEGF antibody – showed superior outcomes (67.2% overall survival (OS) at 12 months vs. 54.6% with sorafenib),20 which led to FDA approval of this combination as first-line systemic treatment for advanced HCC. Second-line treatments include the multikinase inhibitors cabozantinib21 and regorafenib,22 the anti-VEGF antibody ramucirumab,23 the anti-PD1 antibodies nivolumab,24 pembrolizumab25 and the combination of nivolumab with the CTLA-4 inhibitor, ipilimumab.26
With the exception of ramucirumab, for which AFP >400 ng/dl is associated with response, there are no biomarkers to stratify patients with HCC. Thus, emerging novel biomarkers such as circulating tumour DNA (ctDNA) – the tumour-specific component of circulating cell-free DNA (cfDNA) – have garnered substantial attention in the last few years, owing to their potential to address 3 key clinical problems. First, can they be used to detect early HCC at a stage when current surveillance modalities are insensitive and difficult to access? Second, will cfDNA levels obtained before or after surgery be prognostic of long-term outcomes? Lastly, can cfDNA associated with HCC predict response to treatment? In this review, we discuss the most recent developments on the role and utility of circulating cfDNA in the detection and prognostication of HCC.
Liquid biopsy in HCC
Role of cfDNA
Liquid biopsy, the minimally invasive assay of circulating cancer-associated biomarkers such as circulating nucleic acids, circulating tumour cells, miRNAs and exosomes, has several potential clinical applications.27,28 Of these, the analysis of cfDNA is currently the most promising in HCC. Circulating cfDNA refers to fragments of DNA detected in both healthy individuals and patients with cancer.29,30 cfDNA mostly comprises DNA shed from the normal turnover of lymphoid and myeloid cells,31 with ctDNA making up less than 1% of total cfDNA in patients with cancer (Fig. 1).32 Most circulating cfDNA fragments are double-stranded, exist in plasma or serum, and are longer than 167 base pairs.[33], [34], [35] This size approximates the length of DNA wrapped around a single nucleosome, which may protect DNA from degradation by blood nucleases. In contrast, ctDNA fragments, which are released by necrotic or apoptotic tumour cells, are typically shorter than 150 base pairs; these size differences, as well as sequence variation or epigenetic modifications, may be exploited to identify tumour-specific sequences.34
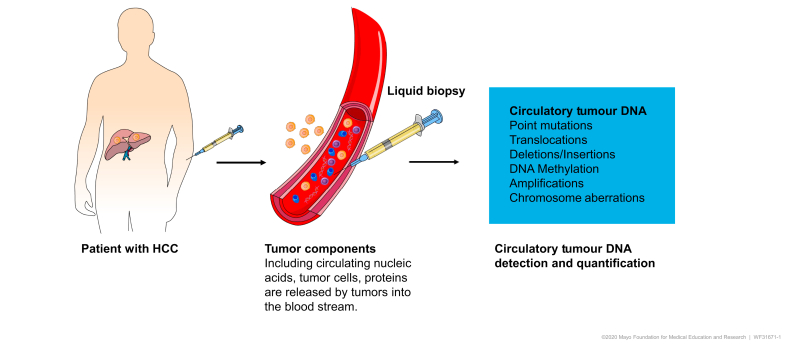
Fig. 1.
Detection and quantification of circulating tumour DNA.
Tumour components including circulatory nucleic acids, tumour cells and proteins are released into the blood stream. Of these, circulating tumour DNA analysis provides information regarding mutations, translocations, deletions/insertions, amplifications and DNA methylation patterns.
Indeed, cancer-specific alterations in ctDNA are measurable by next-generation sequencing (NGS) and targeted PCR-based technologies.36 Early studies demonstrated that ctDNA harbour molecular characteristics known to be present in the genomic DNA of cancers, such as methylation changes[37], [38], [39], [40] and point mutations,[41], [42], [43], [44] which reflect the molecular heterogeneity of a cancer that may be comprised of different tumour clones and metastases. This non-invasive approach involving cfDNA/ctDNA sampling in liquid biopsy is of great interest as it overcomes the limitation of traditional tissue biopsy and temporally reflects the clonal evolution in real time. Potential clinical utilities of cfDNA/ctDNA have been and are being investigated for the detection of HCC,[45], [46], [47], [48] disease monitoring,[49], [50], [51] and prognostication.41,43,52
Storage and detection technology platforms
CtDNA targets must be detected among the background of total cfDNA. Given the short half-life (of between 16 mins and 2.5 h53) and low abundance of ctDNA, it is important to select the right sample collection tube and optimal processing methods to ensure successful DNA isolation. The concentration of cfDNA has been shown to be about 20-fold higher in serum than in matched plasma samples, predominantly as a result of the clotting process in the collection tube.54 Thus, plasma is the preferred biological sample. During handling and processing of whole blood into plasma, lysis of leukocytes can cause enormous contamination of plasma with genomic DNA, reducing if not eliminating, resolution for ctDNA targets. CtDNA is most commonly extracted from peripheral blood plasma; in contrast, processing of whole blood to serum results in shearing and lysis of leukocytes in the clot matrix. DNAases in circulation or sampled whole blood can cause potential loss of global cfDNA from the time of collection to processing to storage to analysis. Specialised tubes such as LBguard (Biomatrica, San Diego CA), Streck (La Vista NE) or Cell-Free DNA Collection (Roche, Basel Switzerland) tubes contain proprietary cfDNA preservation and cell stabilisation buffers for better ctDNA yield and quality.55 The separation of plasma from whole blood requires a 2000 x g centrifugation. The recommended storage temperature is -80oC.
In the last decade, more robust methods with high analytical sensitivity have been developed for ctDNA analysis. These include digital droplet PCR, BEAMing (Beads, Emulsion, Amplification, Magnetic) technology, quantitative allele-specific real-time target and signal amplification and the resulting TELQAS (target enrichment long-probe quantitative amplified signal).[56], [57], [58] These methods allow for more targeted analyses of single nucleotide mutations or methylation changes for example. While the targeted PCR approach is lower cost and has very high sensitivity (mutation to wild-type ratios as low as 0.01%),59 small gene panels will miss mutations that are not selected. Much more costly, untargeted NGS approaches include whole genome sequencing (WGS), whole exome sequencing (WES) and whole genome bisulfite sequencing to screen the genome, exome or methylome for the discovery of known and new aberrations.32,60 A hybrid of these approaches uses NGS after targeted capture of hundreds or even thousands of known allelic or methylated variants.
Early detection of HCC
Early detection of HCC is critical as curative approaches are available when the tumour is small. Despite the current recommendations, standard ultrasound has several disadvantages including suboptimal performance (with sensitivity as low as 42% for lesions smaller than 1 cm), being operator dependent, and involving a cumbersome process for patients. Both CT and MRI perform better for the early detection of HCC. A systematic review including 20 studies reported a pooled sensitivity of 67.5% at 92.5% specificity for CT and 80.6% at 84.8% specificity for MRI.61 For lesions greater than 2 cm, these imaging modalities showed great sensitivity and specificity. However, for small tumours and those that lack arterial-phase hyper-enhancement, which may be up to 40% of HCC, these imaging modalities are limiting in diagnosing HCC. For HCC ≤1 cm in size, detection rates can be as low as 34% and 10% for MRI and CT, respectively.62 Other disadvantages of these tests include cost, access and radiation exposure. Thus, there is a clear need for non-invasive biomarkers that can identify early disease, ideally those patients with lesions <2 cm, and thereby minimise morbidity and mortality associated with late-stage disease. Such measures would also enable the implementation of more efficient and cost-effective surveillance strategies.
Quantitative cfDNA measurement
Early studies investigated the clinical utility of cfDNA concentration as a biomarker for detecting HCC. For example, Iizuka et al. observed an increase in cfDNA concentration in 52 patients with HCV-associated HCC compared to HCV carriers, with an optimal cut-off of 73.0 ng/ml.63 Since then, several studies have reported significantly higher cfDNA concentrations in patients with HCC compared to those with chronic hepatitis and almost 20 times that of healthy controls.[64], [65], [66] Despite these findings, several weaknesses exist with this approach. These studies were carried out in both serum and plasma samples, reflecting the different concentrations of cfDNA. Furthermore, different studies utilised different cut-off values to discriminate high or low cfDNA concentration, suggesting that the level is assay platform dependent. Importantly, quantitative analysis of cfDNA does not provide information about the origin of the tumour, molecular alterations or potential targets.
Recently, a model integrating cfDNA levels with age and AFP reported higher detection capability, with an AUC of 0.98 (95% CI 0.92–1.00) at sensitivity of 87% and specificity of 100%.67 This suggests that quantitative cfDNA analysis may still hold promise when it is combined with other protein or genetic markers for the detection of HCC.
Qualitative cfDNA measurement
Significant research interest in recent years has focused on the molecular characteristics of ctDNA in plasma, including methylation patterns and hotspot mutations. In general, these studies have yielded great detection potential in HCC. Table 1 summarises some of the most recent results and performance of these biomarkers.
Table 1.
Performance of ctDNA for early detection of HCC in selected studies.
| Study | Target | Patients | Sensitivity | Specificity | AUC | Limitations |
|---|---|---|---|---|---|---|
| ctDNA methylation profiling | ||||||
| Wang et al.,81 2020 | 21 DMRs | 148 HCC, 84 healthy controls (training) 112 HCC, 96 healthy controls (validation) |
82.9% | 94% | 0.94 | Healthy controls |
| Yang et al.,83 2020 | 39 DMRs | 140 HCC, 84 healthy controls (diagnostic) 155 HCC, 96 healthy controls, 21 HBV, 34 benign liver disease (validation) |
81% (diagnostic) 75% (validation) |
91% (diagnostic) Validation not reported |
0.93 (diagnostic) 0.90 (validation) |
Low number of at-risk controls |
| Chalasani et al.,110 2020 | HOXA1, EMX1, TSPYL5, B3GALT6, AFP, AFP-L3, and sex | 135 HCC, 302 controls (viral and non-viral) BCLC 0-A: 56% |
71% (early stage) 81% (pooled) |
89% | 0.86 (early stage) 0.91 (pooled) |
Follow-up study of Kisiel et al. |
| Roy et al.86, 2019 | Not reported | 60 HCC, 10 benign liver disease, 30 healthy, 30 other cancer type | 95% | 98% | Not reported | Follow-up study of Xu et al. Small cirrhotic controls, different stages of cancer |
| Kisiel et al.80, 2019 | 6 markers | 95 HCC, 51 cirrhosis, 98 healthy controls BCLC 0-A: 48% |
95% (91% for BCLC 0/A) | 92% | 0.94 | Small cirrhotic controls, small number with early-stage disease |
| Cai et al.,84 2019 | 5-hmC based 32 gene panel | 335 HCC, 263 HBV/cirrhosis, 522 healthy controls (training) BCLC 0-A: 100% 809 HCC, 129 HBV/cirrhosis, 256 healthy controls (validation) BCLC 0-A: 27% |
89.6% (training) 82.7% (validation) |
78.9% (training) 76.4% (validation) |
0.92 (training) 0.88 (validation) |
|
| Oussalah et al.75, 2018 | SEPT9 | 98 HCC, 191 cirrhosis BCLC 0-A: 25% |
81%-97% | 69%-96% | 0.94 (pooled) | Single target |
| Xu et al.,82 2017 |
10 markers |
1098 HCC, 835 healthy controls Stage I: 16% |
83%-86% |
91%-95% |
0.94-0.97 |
Healthy controls |
| ctDNA mutation profiling | ||||||
| Tao et al.,111 2020 | Somatic copy number aberrations | 108 HCC, 101 HBV controls (discovery) BCLC 0-A: 67% 89 HCC, 86 HBV (validation) BCLC 0-A: 100% |
70% (early stage) | 95% | 0.89 (pooled) | Limited to HBV controls, retrospective data |
| Qu et al.,94 2019 | 4 genes (TP53, CTNNB1, AXIN1, TERT) + HBV insertion site, AFP, DCP | 65 HCC, 70 non-HCC (training) 331 at-risk patients (validation) |
85% (training) 100% (validation) |
93% (training) 94% (validation) |
Not reported | Low positive predictive value of 17%, younger healthy control in training cohort |
| Cai et al.,91 2019 | Copy number variants and single nucleotide variants, AFP, AFP-L3, DCP | 34 resected HCC | 100% | Not reported | Not reported | Small sample size |
AFP, alpha-fetoprotein; BCLC, Barcelona Clinic liver cancer; ctDNA, circulating tumour DNA; DMRs, differentially methylated regions; DCP, des-γ-carboxy-prothrombin; HCC, hepatocellular carcinoma.
Analysis of epigenetic changes in ctDNA in HCC
Epigenetic modification, such as DNA methylation, plays a crucial role in regulating gene activity both in normal and cancerous cells.68 While cancer cells exhibit global loss of DNA methylation, hypermethylation at CpG islands and promoters is highly tumour specific and quantifiable.[69], [70], [71], [72] In HCC specifically, inactivation of tumour suppressor genes by aberrant methylation of CpG islands is thought to play an early and important role in the pathogenesis of disease.73,74 Hence, screening for these changes that are highly unique to the tumours may allow for early cancer detection. Indeed, recent studies have identified several diagnostic methylation markers that can discriminate HCC from controls with excellent sensitivity and specificity (Table 1). In 1 study, Oussalal and colleagues identified single-target SEPT9 as a good diagnostic cfDNA methylation marker.75 Among 98 patients with HCC and 191 controls, methylated SEPT9 in plasma DNA yielded high accuracy for the detection of HCC with an AUC of 0.94. This test has received the CE Mark for the detection of liver cancer among patients with cirrhosis in Europe (October 2018).76 A prospective multicentre study to determine its diagnostic performance in a US cohort had completed recruitment as of January 2020 (ClinicalTrials.gov Identifier: NCT03804593) and a larger phase II prospective cross-sectional study assessing the diagnostic accuracy of HCC detection in 440 patients with cirrhosis is ongoing (ClinicalTrials.gov Identifier: NCT03311152). Other promising single hypermethylation candidates include VIM, FBLN1, TFPI2, TGR5, MT1M and MTIG.[77], [78], [79]
Several studies have taken advantage of genome-wide methylome sequencing to identify combination methylation panels for improved detection of HCC.[80], [81], [82], [83], [84], [85] One study utilised methylated CpG tandem amplification and sequencing in a genome-wide detection of hypermethylated CpG islands in patients with HCC.85 In a small phase I pilot study involving 36 patients with HCC, 17 with cirrhosis and 38 healthy controls, the authors identified RGS10, ST8SIA6, RUNX2 and VIM as high-performance markers for detection of small HCC (≤3 cm). The combination achieved a sensitivity of 94% at 89% specificity. Of interest, comparing DNA methylation between matched plasma and tissue samples from 10 patients with HCC, the authors found both cancer and non-cancerous tissues contributed to the elevation of the methylation markers found in the plasma of patients with HCC. In another larger study involving 1,098 patients with HCC and 835 healthy controls, the authors constructed a diagnostic panel of 10 methylated markers using methylation profiles of HCC tumours from The Cancer Genome Atlas in addition to an independent data set from normal blood leukocytes.82 When validated in cfDNA, the model achieved a sensitivity of 83% at 91% specificity (AUC 0.94) in distinguishing patients with HCC from normal healthy controls. Despite the excellent performance, a limitation is that controls were healthy individuals. A recent follow-up study involving 130 individuals (both patients with HCC and controls) reported similar results (sensitivity 95%, specificity 98%)86 leading to FDA breakthrough device designation (September 2019) for early detection of HCC. A clinical trial is ongoing to compare the performance of this panel alone, ultrasound alone or the combination of the methylation panel and ultrasound for the detection of HCC in patients with cirrhosis (ClinicalTrials.gov Identifier: NCT03694600). More recently, another study performed whole methylome discovery with identification of novel methylated DNA markers for HCC detection.80 Among 244 patients with HCC, cirrhosis or healthy controls, a 6-marker cfDNA methylation panel yielded similar sensitivity of 95% at 92% specificity (AUC 0.94). Importantly, the panel was able to detect 75% of patients with stage 0 and 93% with stage A HCC.80 In a follow-up study involving 136 patients with HCC and 401 controls,87 a panel of 3 methylated markers (HOXA1, TSPYL5, B3GALT6) in combination with sex and AFP showed 70% sensitivity and 89% specificity for detection of early-stage HCC. This panel has received FDA breakthrough device designation and further validation studies are ongoing (ClinicalTrials.gov Identifier: NCT03628651).
In summary, the aforementioned studies demonstrate that methylation profiling of plasma DNA has great potential for the detection of HCC; additional large prospective validation studies in cohorts of patients with cirrhosis undergoing active surveillance – who represent the ideal target population – are warranted.
Analysis of mutations in ctDNA in HCC
The detection and analysis of somatic genetic alterations in ctDNA might be particularly useful in diagnosing disease, monitoring treatment response, and identifying mutations associated with treatment resistance. Additionally, ctDNA analysis may overcome the challenge of tumour tissue heterogeneity faced by focal tumour biopsy strategies. With improvements in NGS technology and a better understanding of the mutational landscape of HCC, several recent studies have performed a more comprehensive analysis of ctDNA with improved performance over that of single hotspot interrogation.41,43,50,88,89 In 1 study, exome sequencing showed that 83% of mutations identified in the liver were also detected in cfDNA.89 In another analysis of 30 HCC tissues and corresponding cfDNA using a targeted panel of 46 genes, ctDNA was detected in 63% of patients.88 The sensitivity of ctDNA increased to 87% in patients with large tumours (≥5 cm diameter) or metastatic disease. Not surprisingly, consistent with studies on cfDNA quantification and methylation, the ability to detect mutations in ctDNA is associated with tumour burden, vascular invasion and extrahepatic metastasis.43,89,90 Of interest, 81% of mutations detected in cfDNA were independently detected in the corresponding tumour biopsy.88 In agreement with other studies, the copy number profiles of ctDNA reflect the biology of the matched primary tumour.33,43,59
Several studies have shown that ctDNA mutation profiling can be used as a tool to monitor disease dynamics including response to treatment and disease progression.38,59,89 These studies showed that following resection, ctDNA levels dropped or disappeared completely and rose again prior to disease progression. A more recent study targeting 574 genes in tumour tissues of 3 patients with HCC revealed that 98%–99% of identified subclonal mutations were captured in ctDNA.49 Furthermore, the level of subclonal mutations changed in correlation with the patient’s tumour burden, with a lower fraction of mutated alleles detected after resection and a higher mutational frequency observed during recurrence. In a follow-up study of 34 patients with HCC, the authors performed WES to a median depth of 152x to identify specific single nucleotide variants (SNVs) and copy number variants (CNVs) in tumour tissue and peritumoral tissues.91 Leveraging individual findings, custom-made panels were designed to capture these mutations in patient’s plasma at a median depth of 7,204x. The threshold levels of SNVs and CNVs in ctDNA were detected in all preoperative patients with HCC and in 95% of patients at the time of tumour recurrence (compared to 49%, 45% and 77% for AFP, AFP-L3 and des-γ-carboxy-prothrombin [DCP], respectively). Serial measurement of ctDNA postoperatively identified 59% of patients with recurrence within 1 year, suggesting the feasibility of monitoring for minimal residual disease.
Furthermore, ctDNA carries genetic information integrated from the entire tumour mass, circumventing the challenges posed by intratumoural heterogeneity when only focal tumour biopsies are obtained.59,92 This concept was explored using shotgun massively parallel sequencing of plasma DNA in 4 patients with HCC before surgery and comparing it to multiregional sequencing of tumour tissue. The results revealed that up to 94% of tumour-associated SNVs were detected in cfDNA.59 Another study performed WES and targeted deep sequencing of 32 multiregional HCC tissue specimens from 5 patients, highlighting the challenges with mutation profiling of ctDNA.92 WES of ctDNA revealed only 18% of the mutations detected in tissue. When targeted deep sequencing was applied, detection increased to 84%. In this small study, the authors demonstrated that cfDNA captured most of mutations between tumour regions; however, this required a higher depth of sequencing to a median sequencing depth of 1,807x vs. 226x. Finally, a more recent study evaluating the concordance of ctDNA and tissue using a targeted panel of 8 genes among 51 patients with HCC found mutations in ctDNA from only 35% of patients.93 In patients with matched tissue DNA, 71% of mutations found in tissue were not detected in matched ctDNA. Thus, this approach lacked sensitivity.
A more exciting recent development in detecting HCC is the combination of DNA mutations with cancer-associated proteins. One study combining a panel of 4 genes (TP53, CTNNB1, AXIN1, TERT), AFP and DCP discriminated 65 patients with HCC from 70 without HCC, with 85% sensitivity at 93% specificity in the training cohort.94 The test yielded a sensitivity of 100% and specificity of 94% in the validation cohort of 331 at-risk patients; however, the positive predictive value was only 17%, reflecting the healthy younger training cohort in this study. In another study, Cohen and colleagues combined circulating proteins with NGS and detected early-stage cancer with sensitivities ranging from 69% to 98% for 5 cancer types.95 In liver cancer in particular, the assay achieved a sensitivity of 95% with over 99% specificity.
In summary, the variability in the proportion of patients with HCC and detectable ctDNA reflects not only cohort composition and methodologies of detection but also clinical characteristics of the disease. Nevertheless, these studies also showcase the feasibility and potential applicability of cfDNA as a diagnostic marker.
Prognostic value of ctDNA in HCC
Outside of HCC detection, ctDNA can also play an important role in prognostication. Earlier studies showed shorter disease-free survival and overall survival were associated with higher cfDNA concentration,52,65 higher levels of cfDNA methylation markers,39,40 and specific hotspot mutations.43,89 In a recent study, analysis of 155 patients with HCC undergoing surgical resection showed that promoter methylation of insulin-like growth factor-binding protein 7 was associated with early tumour recurrence and decreased overall survival after hepatectomy.96 In 1 of the largest studies to date, including over 1,000 patients (Table 2), Xu and colleagues identified 8 methylation markers which independently predicted worse overall survival in both training and validation cohorts.82
Table 2.
Performance of ctDNA for disease monitoring or prognostication in selected studies.
| Study | Target | Patients | Performance | Limitations |
|---|---|---|---|---|
| ctDNA mutation profiling | ||||
| Kim et al.,97 2020 | 2,924 SNVs in 69 genes | 107 HCC | MLH1 is associated with poor overall survival | Single MLH1 target |
| Zhou et al.,98 2020 | 1,021 gene panel (target not reported) | 97 HCC, resected | Associated with shorter disease-free survival | Single liquid biopsy |
| Alunni-Fabbroni et al.,90 2019 | 597 gene panel | 13 HCC (SORAMIC trial) | cfDNA levels associated with presence of metastases and survival | Small sample size |
| Oh et al.,99 2019 | VEGFA, copy number alteration | 151 HCC, 14 healthy controls | Higher cfDNA associated with shorter time to progression (sorafenib), shorter overall survival | Exploratory study |
| Cai et al.,91 2019 |
CNVs and SNVs, AFP, AFP-L3, DCP |
34 HCC, resected |
High SNV and CNV correlated with shorter relapse-free survival and overall survival |
Small sample size |
| ctDNA methylation profiling | ||||
| Xu et al.,82 2017 | 8 markers | 1,098 HCC, 835 healthy controls | Combined prognosis score predicted worse overall survival | Short follow-up |
AFP, alpha fetoprotein; cfaDNA, cell-free DNA; CNV, copy number variants; ctDNA, circulating tumour DNA; DCP, des-γ-carboxy-prothrombin; HCC, hepatocellular carcinoma; SNV, single nucleotide variants.
Similarly, recent studies targeting specific mutations in cfDNA reflect intratumoral heterogeneity and predict poor prognosis. Kim and colleagues analysed 2,924 SNVs in 69 genes from 61 patients and validated SNVs in MLH1, STK11, PTEN and CTNNB1 by digital droplet PCR.97 Of these, MLH1 was found in both ctDNA and tumour tissue and was associated with shorter overall survival. In another study, analysis of >1,000 genes from 97 patients undergoing resection revealed that the presence of ctDNA 7 days after surgery was an independent predictor of poor prognosis.98 Twenty-one patients had at least 1 mutation and all of them recurred. A selection of recent studies have reported similar results for other genes (Table 2).90,91,99
Current status and future considerations
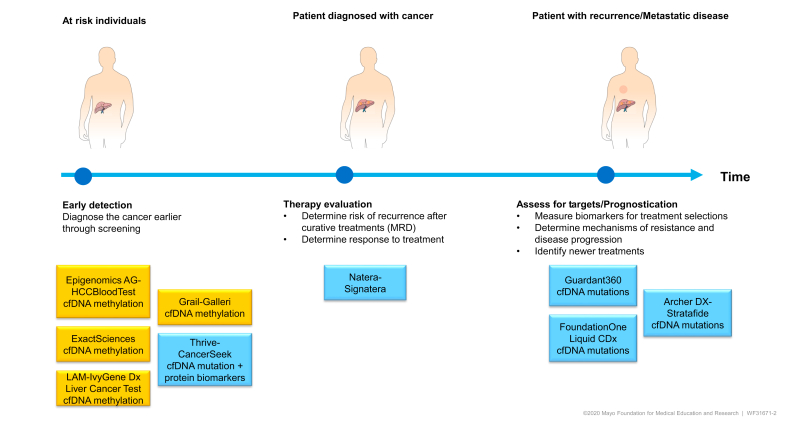
Precision oncology has undoubtedly transformed the clinical management of patients with cancer. With the improvements in NGS technologies accompanied by the increasing understanding of the molecular pathogenesis of HCC, ctDNA detection and characterisation carry immense potential for clinical application (Fig. 2). From early tumour detection to prognostication and therapy evaluation, several tests are in advanced stages of clinical development. In early tumour detection, 3 companies have received breakthrough device designation/CE Mark for their individual tests (ExactSciences,100 Laboratory for Advanced Medicine,82 and Epigenomics AG75). Another example includes the multi-methylation target panel, Galleri™ (GRAIL, Menlo Park CA). GRAIL launched the Circulating Cell-free Genome Atlas Study (CCGA), a prospective, observational, longitudinal clinical trial (NCT02889978) designed to determine detection and localisation of tumour origin in 50 cancer types by combining genome-wide cfDNA sequencing and machine learning. Recent updates from a sub-study cohort of >6,000 participants (>2,000 patients with cancer from >50 cancer types and >4,000 individuals without cancer) showed increasing sensitivity with disease stage (39% in stage I to 92% in stage IV), with tissue of origin localisation predicted in 96% of samples with 93% accuracy.101 GRAIL received breakthrough device designation in May 2019 and is planning to launch their product as a lab developed test in 2021. Another recent update included the first study of its kind where the authors combined the CancerSeek blood test (including 16 gene mutations and 9 protein biomarkers) with PET/CT to detect cancer in over 10,000 women without a history of cancer or symptoms.102 Twenty-six cancers were detected with sensitivity of 27%, specificity of 99% and positive predictive value of 19%. A new generation of this test has shown higher sensitivity without compromising specificity.95 This study showed the feasibility and safety of administering a cancer screening blood test with subsequent confirmation tests and imaging in a large prospective cohort. Only 38 women received false-positive test results and most of these women had non-invasive or minimally invasive testing. The design and application of this study illustrates the profound and near-term potential of cfDNA for the early detection of multiple cancers.
Fig. 2.
Clinical applications of cfDNA.
Select examples of clinical tests having received breakthrough device designation or FDA approval. Colour denotes the technology used. cfDNA, cell-free DNA.
While the evaluation of cfDNA for early detection is a promising strategy, several outstanding issues warrant discussion. First, the lack of standardised protocols for preanalytical sample preparation and ctDNA purification, and different platforms for analysis, have resulted in significant variability in test sensitivity and specificity and hindered biomarker development in HCC. Standardisation of these factors would ensure consistency of results. The most promising approach, which is currently undergoing clinical validation, is DNA methylation profiling (Fig. 2). Previous work suggests that DNA methylation profiling is more broadly informative than mutation-based strategies which require WGS103 and targeted sequencing of much larger variants including CNVs and SNVs.104 Alterations in DNA methylation appear to be more prevalent than driver mutation sites and also provide an epigenetic memory of tissue of origin. They are therefore effective molecular markers for tumour detection.101 Exploiting combinations of DNA profiling with protein biomarkers have shown promising results as well and it is likely that multi-omics approaches will be developed for clinical use.
Second, while preclinical exploratory studies have generated significant proof-of-concept evidence of ctDNA as a surveillance blood test, large well-controlled longitudinal studies are still lacking. Of the 5 phases of biomarker development for early detection articulated by the Early Detection Research Network105 and more recently by the International Liver Cancer Association,106 there are few phase II clinical assay development studies and even fewer phase III studies that include independent prospective validation cohorts, particularly of patients with cirrhosis undergoing surveillance. Importantly, the detection of ctDNA mutations and methylations in advanced-stage cancer have been successful, however, current studies included low numbers of Barcelona Clinic liver cancer stage 0 patients (Table 1), which could lead to overestimation of biomarker performance for the early detection of liver cancer. Establishing acceptable sensitivity and specificity in this target population is crucial to avoid high rates of false negative/positive results. This lack of evidence and the off-target population are major impediments to biomarker development and clinical translation.107 Given the increasing interest and extensive research in identifying biomarkers for cancer detection and management, the International Liver Cancer Association also provided a framework on best practices in study design and interpretation of biomarker studies.106 The success of biomarker development will not only involve the right study design, including the target population, and interpretation but also investment in phase II studies and beyond by key players including government, industry and public-private partnerships.108 This evidence will be of paramount importance for the translation of these biomarkers into practice.
Finally, biomarkers must show clinical utility, demonstrating that they can improve health outcomes relative to the existing standard alternative and that the results can guide subsequent clinical management.109 The clinical utility of a diagnostic test is often measured as the expected number of life years gained with adjustment for the quality of those years.109 More evidence in the form of randomised clinical trials or decision analysis models of the clinical utility of cfDNA is needed. Importantly, there is substantial uncertainty in estimating the cost-effectiveness of these biomarkers in the detection of HCC.
In conclusion, the potential applicability of cfDNA as a biomarker for disease detection and prognostication in HCC has been clearly demonstrated. Its role in precision oncology is promising and will likely enhance cancer management for many patients in the near future.
Financial support
LRR was supported by the Mayo Clinic Hepatobiliary SPORE (P50 CA210964) and the Mayo Clinic Center for Clinical and Translational Science (UL1 TR002377).
Authors’ contributions
All authors contributed to the writing and review of this manuscript.
Conflict of interest
Dr. Kisiel is an inventor of Mayo Clinic intellectual property licensed to Exact Sciences (Madison WI) and may receive royalties paid to Mayo Clinic under a sponsored research contract.
Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2021.100304.
Supplementary data
The following is the supplementary data to this article:
References
- 1.Akinyemiju T., Abera S., Ahmed M., Alam N., Alemayohu M.A., Allen C. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level. JAMA Oncol. 2017;98121:1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henley S.J., Ward E.M., Scott S., Ma J., Anderson R.N., Firth A.U. Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer. 2020;126:2225–2249. doi: 10.1002/cncr.32802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White D.L., Thrift A.P., Kanwal F., Davila J., El-Serag H.B. Incidence of hepatocellular carcinoma in all 50 United States, from 2000 through 2012. Gastroenterology. 2017;152 doi: 10.1053/j.gastro.2016.11.020. 812-820.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Serag H.B. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 5.Singal A.G., Pillai A., Tiro J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: a meta-analysis. Plos Med. 2014;11 doi: 10.1371/journal.pmed.1001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B.H., Yang B.H., Tang Z.Y. Randomized controlled trial of screening for hepatocellular carcinoma. J Canc Res Clin Oncol. 2004;130:417–422. doi: 10.1007/s00432-004-0552-0. [DOI] [PubMed] [Google Scholar]
- 7.EASL Clinical Practice Guidelines Management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 8.Marrero J.A., Kulik L.M., Sirlin C.B., Zhu A.X., Finn R.S., Abecassis M.M. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American association for the study of liver diseases. Hepatology. 2018;68:723–750. doi: 10.1002/hep.29913. [DOI] [PubMed] [Google Scholar]
- 9.Tzartzeva K., Obi J., Rich N.E., Parikh N.D., Marrero J.A., Yopp A. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154:1706–1718.e1. doi: 10.1053/j.gastro.2018.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolf E., Rich N.E., Marrero J.A., Parikh N.D., Singal A.G. Use of hepatocellular carcinoma surveillance in patients with cirrhosis: a systematic review and meta-analysis. Hepatology. 2020 doi: 10.1002/hep.31309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazzaferro V., Regalia E., Doci R., Andreola S., Pulvirenti A., Bozzetti F. Carcinomas in patients with cirrhosis. N Engl J Med. 1996;334:693–699. doi: 10.1056/NEJM199603143341104. [DOI] [PubMed] [Google Scholar]
- 12.Yao F.Y., Ferrell L., Bass N.M., Watson J.J., Bacchetti P., Venook A. Liver transplantation for hepatocellular carcinoma: expansion of the tumor size limits does not adversely impact survival. Hepatology. 2001;33:1394–1403. doi: 10.1053/jhep.2001.24563. [DOI] [PubMed] [Google Scholar]
- 13.Chapman W.C., Klintmalm G., Hemming A., Vachharajani N., Majella Doyle M.B., Dematteo R. Surgical treatment of hepatocellular carcinoma in North America: can hepatic resection still be justified? J Am Coll Surgeons. 2015;220:628–637. doi: 10.1016/j.jamcollsurg.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 14.Li Y., Xia Y., Li J., Wu D., Wan X., Wang K. Prognostic nomograms for pre- and postoperative predictions of long-term survival for patients who underwent liver resection for huge hepatocellular carcinoma. J Am Coll Surgeons. 2015;221:962–974.e4. doi: 10.1016/j.jamcollsurg.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 15.She W.H., Chok K.S.H., Ang S.F., Ng E.S.H., Li H., Ong Y.H. The Singapore Liver Cancer Recurrence (SLICER) Score for relapse prediction in patients with surgically resected hepatocellular carcinoma. Ann Surg. 2015;261 doi: 10.1371/journal.pone.0118658. 962-974.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shim J.H., Jun M.J., Han S., Lee Y.J., Lee S.G., Kim K.M. Prognostic nomograms for prediction of recurrence and survival after curative liver resection for hepatocellular carcinoma. Ann Surg. 2015;261:939–946. doi: 10.1097/SLA.0000000000000747. [DOI] [PubMed] [Google Scholar]
- 17.Yang P., Qiu J., Li J., Wu D., Wan X., Lau W.Y. Nomograms for pre-and postoperative prediction of long-term survival for patients who under went hepatectomy for multiple hepatocellular carcinomas. Ann Surg. 2016;263:778–786. doi: 10.1097/SLA.0000000000001339. [DOI] [PubMed] [Google Scholar]
- 18.Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 19.Kudo M., Finn R.S., Qin S., Han K.H., Ikeda K., Piscaglia F. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. The Lancet. 2018;391:1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 20.Finn R.S., Qin S., Ikeda M., Galle P.R., Ducreux M., Kim T.Y. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382:1894–1905. doi: 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- 21.Abou-Alfa G.K., Meyer T., Cheng A.L., El-Khoueiry A.B., Rimassa L., Ryoo B.Y. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N Engl J Med. 2018;379:54–63. doi: 10.1056/NEJMoa1717002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruix J., Qin S., Merle P., Granito A., Huang Y.H., Bodoky G. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 23.Zhu A.X., Kang Y.K., Yen C.J., Finn R.S., Galle P.R., Llovet J.M. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20:282–296. doi: 10.1016/S1470-2045(18)30937-9. [DOI] [PubMed] [Google Scholar]
- 24.El-Khoueiry A.B., Sangro B., Yau T., Crocenzi T.S., Kudo M., Hsu C. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu A.X., Finn R.S., Edeline J., Cattan S., Ogasawara S., Palmer D. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19:940–952. doi: 10.1016/S1470-2045(18)30351-6. [DOI] [PubMed] [Google Scholar]
- 26.Yau T., Kang Y.-K., Kim T.-Y., El-Khoueiry A.B., Santoro A., Sangro B. Efficacy and safety of nivolumab plus ipilimumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib: the CheckMate 040 randomized clinical trial. JAMA Oncol. 2020;6 doi: 10.1001/jamaoncol.2020.4564. e204564-e204564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felden J.V., Craig A.J., Villanueva A. Role of circulating tumor DNA to help decision-making in hepatocellular carcinoma. Oncoscience. 2018;5 doi: 10.18632/oncoscience.446. 1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J., Han X., Yu X., Xu Z., Yang G., Liu B. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J Exp Clin Canc Res. 2018;37:1–13. doi: 10.1186/s13046-018-0893-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandel P., Metais P. Nuclear acids in human blood plasma. C R Seances Soc Biol Fil. 1948;142:241–243. [PubMed] [Google Scholar]
- 30.Sorenson G.D., Pribish D.M., Valone F.H., Memoli V.A., Bzik D.J., Yao S.L. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Canc Epidemiol Biomark Prev. 1994;3:67–71. [PubMed] [Google Scholar]
- 31.Snyder M.W., Kircher M., Hill A.J., Daza R.M., Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164:57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman A.M., Bratman S.V., To J., Wynne J.F., Eclov N.C., Modlin L.A. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang P., Chan C.W., Chan K.C., Cheng S.H., Wong J., Wong V.W. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A. 2015;112:E1317–E1325. doi: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouliere F., Chandrananda D., Piskorz A.M., Moore E.K., Morris J., Ahlborn L.B. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Underhill H.R., Kitzman J.O., Hellwig S., Welker N.C., Daza R., Baker D.N. Fragment length of circulating tumor DNA. Plos Genet. 2016;12 doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diaz L.A., Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balgkouranidou I., Chimonidou M., Milaki G., Tsarouxa E.G., Kakolyris S., Welch D.R. Breast cancer metastasis suppressor-1 promoter methylation in cell-free DNA provides prognostic information in non-small cell lung cancer. Br J Canc. 2014;110:2054–2062. doi: 10.1038/bjc.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan K.C., Jiang P., Chan C.W., Sun K., Wong J., Hui E.P. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A. 2013;110:18761–18768. doi: 10.1073/pnas.1313995110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan K.C., Lai P.B., Mok T.S., Chan H.L., Ding C., Yeung S.W. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin Chem. 2008;54:1528–1536. doi: 10.1373/clinchem.2008.104653. [DOI] [PubMed] [Google Scholar]
- 40.Wong I.H., Lo Y.M., Zhang J., Liew C.T., Ng M.H., Wong N. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Canc Res. 1999;59:71–73. [PubMed] [Google Scholar]
- 41.Huang A., Zhang X., Zhou S.L., Cao Y., Huang X.W., Fan J. Detecting circulating tumor DNA in hepatocellular carcinoma patients using droplet digital PCR is feasible and reflects intratumoral heterogeneity. J Canc. 2016;7:1907–1914. doi: 10.7150/jca.15823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao J., Niu W., Wang Y., Baggerly K., Ye Y., Wu X. Prevalence of aflatoxin-associated TP53R249S mutation in hepatocellular carcinoma in hispanics in south Texas. Canc Prev Res (Phila) 2018;11:103–112. doi: 10.1158/1940-6207.CAPR-17-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao W., Yang H., Xu H., Wang Y., Ge P., Ren J. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget. 2016;7:40481–40490. doi: 10.18632/oncotarget.9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marchio A., Amougou Atsama M., Béré A., Komas N.P., Noah Noah D., Atangana P.J.A. Droplet digital PCR detects high rate of TP53 R249S mutants in cell-free DNA of middle African patients with hepatocellular carcinoma. Clin Exp Med. 2018;18:421–431. doi: 10.1007/s10238-018-0502-9. [DOI] [PubMed] [Google Scholar]
- 45.Huang Z.H., Hu Y., Hua D., Wu Y.Y., Song M.X., Cheng Z.H. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol. 2011;91:702–707. doi: 10.1016/j.yexmp.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Lu C.Y., Chen S.Y., Peng H.L., Kan P.Y., Chang W.C., Yen C.J. Cell-free methylation markers with diagnostic and prognostic potential in hepatocellular carcinoma. Oncotarget. 2017;8:6406–6418. doi: 10.18632/oncotarget.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeo W., Wong N., Wong W.L., Lai P.B., Zhong S., Johnson P.J. High frequency of promoter hypermethylation of RASSF1A in tumor and plasma of patients with hepatocellular carcinoma. Liver Int. 2005;25:266–272. doi: 10.1111/j.1478-3231.2005.01084.x. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y.J., Wu H.C., Shen J., Ahsan H., Tsai W.Y., Yang H.I. Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clin Canc Res. 2007;13:2378–2384. doi: 10.1158/1078-0432.CCR-06-1900. [DOI] [PubMed] [Google Scholar]
- 49.Cai Z.X., Chen G., Zeng Y.Y., Dong X.Q., Lin M.J., Huang X.H. Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. Int J Canc. 2017;141:977–985. doi: 10.1002/ijc.30798. [DOI] [PubMed] [Google Scholar]
- 50.Ikeda S., Tsigelny I.F., Skjevik Å A., Kono Y., Mendler M., Kuo A. Next-generation sequencing of circulating tumor DNA reveals frequent alterations in advanced hepatocellular carcinoma. Oncologist. 2018;23:586–593. doi: 10.1634/theoncologist.2017-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong I.H.N., Zhang J., Lai P.B.S., Lau W.Y., Dennis Lo Y.M. Quantitative analysis of tumor-derived methylated <strong><em>p16INK4a</em></strong> sequences in plasma, serum, and blood cells of hepatocellular carcinoma patients. Clin Canc Res. 2003;9:1047–1052. [PubMed] [Google Scholar]
- 52.Ren N., Qin L.X., Tu H., Liu Y.K., Zhang B.H., Tang Z.Y. The prognostic value of circulating plasma DNA level and its allelic imbalance on chromosome 8p in patients with hepatocellular carcinoma. J Canc Res Clin Oncol. 2006;132:399–407. doi: 10.1007/s00432-005-0049-5. [DOI] [PubMed] [Google Scholar]
- 53.Diehl F., Schmidt K., Choti M.A., Romans K., Goodman S., Li M. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee T.H., Montalvo L., Chrebtow V., Busch M.P. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion. 2001;41:276–282. doi: 10.1046/j.1537-2995.2001.41020276.x. [DOI] [PubMed] [Google Scholar]
- 55.Browne C.D., Mattmann M.E., Wycoco M.J., Chen S.N., Ravichandran R., Desharnais J. Abstract 2758: comparison of cell-free DNA blood collection tubes. Canc Res. 2017;77 2758-2758. [Google Scholar]
- 56.Diehl F., Li M., He Y., Kinzler K.W., Vogelstein B., Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3:551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- 57.Imperiale T.F., Ransohoff D.F., Itzkowitz S.H., Levin T.R., Lavin P., Lidgard G.P. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 58.Taly V., Pekin D., Benhaim L., Kotsopoulos S.K., Le Corre D., Li X. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59:1722–1731. doi: 10.1373/clinchem.2013.206359. [DOI] [PubMed] [Google Scholar]
- 59.Chan K.C., Jiang P., Zheng Y.W., Liao G.J., Sun H., Wong J. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 60.Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Colli A., Fraquelli M., Casazza G., Massironi S., Colucci A., Conte D. Accuracy of ultrasonography, spiral CT, magnetic resonance, and alpha-fetoprotein in diagnosing hepatocellular carcinoma: a systematic review. Am J Gastroenterol. 2006;101:513–523. doi: 10.1111/j.1572-0241.2006.00467.x. [DOI] [PubMed] [Google Scholar]
- 62.Choi J.Y., Lee J.M., Sirlin C.B. CT and MR imaging diagnosis and staging of hepatocellular carcinoma: part I. Development, growth, and spread: key pathologic and imaging aspects. Radiology. 2014;272:635–654. doi: 10.1148/radiol.14132361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iizuka N., Sakaida I., Moribe T., Fujita N., Miura T., Stark M. Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis C virus-associated hepatocellular carcinoma. Anticanc Res. 2006;26:4713–4719. [PubMed] [Google Scholar]
- 64.Huang Z., Hua D., Hu Y., Cheng Z., Zhou X., Xie Q. Quantitation of plasma circulating DNA using quantitative PCR for the detection of hepatocellular carcinoma. Pathol Oncol Res. 2012;18:271–276. doi: 10.1007/s12253-011-9438-z. [DOI] [PubMed] [Google Scholar]
- 65.Tokuhisa Y., Iizuka N., Sakaida I., Moribe T., Fujita N., Miura T. Circulating cell-free DNA as a predictive marker for distant metastasis of hepatitis C virus-related hepatocellular carcinoma. Br J Canc. 2007;97:1399–1403. doi: 10.1038/sj.bjc.6604034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Y.J., Chen H., Huang P., Li C.H., Dong Z.H., Hou Y.L. Quantification of plasma hTERT DNA in hepatocellular carcinoma patients by quantitative fluorescent polymerase chain reaction. Clin Invest Med. 2011;34:E238. doi: 10.25011/cim.v34i4.15366. [DOI] [PubMed] [Google Scholar]
- 67.Yan L., Chen Y., Zhou J., Zhao H., Zhang H., Wang G. Diagnostic value of circulating cell-free DNA levels for hepatocellular carcinoma. Int J Infect Dis. 2018;67:92–97. doi: 10.1016/j.ijid.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 68.Portela A., Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 69.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 70.Costello J.F., Frühwald M.C., Smiraglia D.J., Rush L.J., Robertson G.P., Gao X. Aberrant CpG-island methylation has non-random and tumour-type–specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 71.Esteller M., Corn P.G., Baylin S.B., Herman J.G. A gene hypermethylation profile of human cancer. Canc Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 72.Paz M.F., Fraga M.F., Avila S., Guo M., Pollan M., Herman J.G. A systematic profile of DNA methylation in human cancer cell lines. Canc Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- 73.Yang B., Guo M., Herman J.G., Clark D.P. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol. 2003;163:1101–1107. doi: 10.1016/S0002-9440(10)63469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhong S., Tang M.W., Yeo W., Liu C., Lo Y.M., Johnson P.J. Silencing of GSTP1 gene by CpG island DNA hypermethylation in HBV-associated hepatocellular carcinomas. Clin Canc Res. 2002;8:1087–1092. [PubMed] [Google Scholar]
- 75.Oussalah A., Rischer S., Bensenane M., Conroy G., Filhine-Tresarrieu P., Debard R. Plasma mSEPT9: a novel circulating cell-free DNA-based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine. 2018;30:138–147. doi: 10.1016/j.ebiom.2018.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Epigenomics. Epigenomics AG obtains CE Mark for Liquid Biopsy Test for Liver Cancer Detection.https://www.epigenomics.com/epigenomics-ag-obtains-ce-mark-for-liquid-biopsy-test-for-liver-cancer-detection-2/ [Access Jan 2 2021].
- 77.Han L.Y., Fan Y.C., Mu N.N., Gao S., Li F., Ji X.F. Aberrant DNA methylation of G-protein-coupled bile acid receptor Gpbar1 (TGR5) is a potential biomarker for hepatitis B Virus associated hepatocellular carcinoma. Int J Med Sci. 2014;11:164–171. doi: 10.7150/ijms.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holmila R., Sklias A., Muller D.C., Esposti D.D., Guilloreau P., McKay J. Targeted deep sequencing of plasma circulating cell-free DNA reveals Vimentin and Fibulin 1 as potential epigenetic biomarkers for hepatocellular carcinoma. PloS One. 2017;12:1–15. doi: 10.1371/journal.pone.0174265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ji X.F., Fan Y.C., Gao S., Yang Y., Zhang J.J., Wang K. MT1M and MT1G promoter methylation as biomarkers for hepatocellular carcinoma. World J Gastroenterol. 2014;20:4723–4729. doi: 10.3748/wjg.v20.i16.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kisiel J.B., Dukek B.A., VSRK R., Ghoz H.M., Yab T.C., Berger C.K. Hepatocellular carcinoma detection by plasma methylated DNA: discovery, phase I pilot, and phase II clinical validation. Hepatology. 2019;69:1180–1192. doi: 10.1002/hep.30244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Y., Wang Y., Huang A., Jiang R., Zheng J., Li Z. Abstract 782: the genetic and epigenetic abnormalities of plasma cfDNA as liquid biopsy biomarkers to diagnose hepatocellular carcinoma. Canc Res. 2020;80 782-782. [Google Scholar]
- 82.Xu R.H., Wei W., Krawczyk M., Wang W., Luo H., Flagg K. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16:1155–1162. doi: 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
- 83.Yang X.-R., Huang A., Wang Y., Peng J., Jiang R., Li Z. Genome-wide plasma cell-free DNA methylation profiling to identify high-performing biomarkers for early detection of hepatocellular carcinoma. J Clin Oncol. 2020;38 4600-4600. [Google Scholar]
- 84.Cai J., Chen L., Zhang Z., Zhang X., Lu X., Liu W. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut. 2019;68:2195–2205. doi: 10.1136/gutjnl-2019-318882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wen L., Li J., Guo H., Liu X., Zheng S., Zhang D. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Res. 2015;25:1250–1264. doi: 10.1038/cr.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roy D., Taggart D., Zheng L., Liu D., Li G., Li M. Abstract 837: circulating cell-free DNA methylation assay: towards early detection of multiple cancer types. Canc Res. 2019;79 837-837. [Google Scholar]
- 87.Chalasani N.P., Bhattacharya A., Book A., Neis B., Xiong K., Ramasubramanian T. Algorithm for blood-based panel of methylated DNA and protein markers to detect early-stage hepatocellular carcinoma with high specificity. J Clin Oncol. 2020;38 4577-4577. [Google Scholar]
- 88.Ng C.K.Y., Di Costanzo G.G., Tosti N., Paradiso V., Coto-Llerena M., Roscigno G. Genetic profiling using plasma-derived cell-free DNA in therapy-naïve hepatocellular carcinoma patients: a pilot study. Ann Oncol. 2018;29:1286–1291. doi: 10.1093/annonc/mdy083. [DOI] [PubMed] [Google Scholar]
- 89.Ono A., Fujimoto A., Yamamoto Y., Akamatsu S., Hiraga N., Imamura M. Circulating tumor DNA analysis for liver cancers and its usefulness as a liquid biopsy. Cell Mol Gastroenterol Hepatol. 2015;1:516–534. doi: 10.1016/j.jcmgh.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alunni-Fabbroni M., Rönsch K., Huber T., Cyran C.C., Seidensticker M., Mayerle J. Circulating DNA as prognostic biomarker in patients with advanced hepatocellular carcinoma: a translational exploratory study from the SORAMIC trial. J Translational Med. 2019;17:328. doi: 10.1186/s12967-019-2079-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cai Z., Chen G., Zeng Y., Dong X., Li Z., Huang Y. Comprehensive liquid profiling of circulating tumor DNA and protein biomarkers in long-term follow-up patients with hepatocellular carcinoma. Clin Canc Res. 2019;25:5284–5294. doi: 10.1158/1078-0432.CCR-18-3477. [DOI] [PubMed] [Google Scholar]
- 92.Huang A., Zhao X., Yang X.R., Li F.Q., Zhou X.L., Wu K. Circumventing intratumoral heterogeneity to identify potential therapeutic targets in hepatocellular carcinoma. J Hepatol. 2017;67:293–301. doi: 10.1016/j.jhep.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 93.Howell J., Atkinson S.R., Pinato D.J., Knapp S., Ward C., Minisini R. Identification of mutations in circulating cell-free tumour DNA as a biomarker in hepatocellular carcinoma. Eur J Canc. 2019;116:56–66. doi: 10.1016/j.ejca.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 94.Qu C., Wang Y., Wang P., Chen K., Wang M., Zeng H. Detection of early-stage hepatocellular carcinoma in asymptomatic HBsAg-seropositive individuals by liquid biopsy. Proc Natl Acad Sci U S A. 2019;116:6308–6312. doi: 10.1073/pnas.1819799116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li F., Qiao C.Y., Gao S., Fan Y.C., Chen L.Y., Wang K. Circulating cell-free DNA of methylated insulin-like growth factor-binding protein 7 predicts a poor prognosis in hepatitis B virus-associated hepatocellular carcinoma after hepatectomy. Free Radic Res. 2018;52:455–464. doi: 10.1080/10715762.2018.1443448. [DOI] [PubMed] [Google Scholar]
- 97.Kim S.S., Eun J.W., Choi J.-H., Woo H.G., Cho H.J., Ahn H.R. MLH1 single-nucleotide variant in circulating tumor DNA predicts overall survival of patients with hepatocellular carcinoma. Scientific Rep. 2020;10:17862. doi: 10.1038/s41598-020-74494-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou L., Xu Y., Wang D., Ye K., Xiao L., Long G. Perioperative circulating tumor DNA analysis to predict patient prognosis in liver cancer. J Clin Oncol. 2020;38 4593-4593. [Google Scholar]
- 99.Oh C.R., Kong S.Y., Im H.S., Kim H.J., Kim M.K., Yoon K.A. Genome-wide copy number alteration and VEGFA amplification of circulating cell-free DNA as a biomarker in advanced hepatocellular carcinoma patients treated with Sorafenib. BMC Cancer. 2019;19:292. doi: 10.1186/s12885-019-5483-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chalasani N.P., Ramasubramanian T., Bruinsma J.J., Allawi H.T., Olson M., Roberts L.R. American Association for The Study of Liver Diseases. 2019. Combined methylated DNA and protein markers: an accurate blood based test for early stage detection of hepatocellular carcinoma. November 2019; Boston MA: Abstract nr109. [Google Scholar]
- 101.Liu M.C., Oxnard G.R., Klein E.A., Swanton C., Seiden M.V., Liu M.C. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31:745–759. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lennon A.M., Buchanan A.H., Kinde I., Warren A., Honushefsky A., Cohain A.T. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science. 2020;369 doi: 10.1126/science.abb9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oxnard G.R., Klein E.A., Seiden M., Hubbell E., Venn O., Jamshidi A. Simultaneous multi-cancer detection and tissue of origin (TOO) localization using targeted bisulfite sequencing of plasma cell-free DNA (cfDNA) J Glob Oncol. 2019;5 44-44. [Google Scholar]
- 104.Liu M.C., Klein E., Hubbell E., Maddala T., Aravanis A.M., Beausang J.F. 50O - plasma cell-free DNA (cfDNA) assays for early multi-cancer detection: the circulating cell-free genome atlas (CCGA) study. Ann Oncol. 2018;29 viii14. [Google Scholar]
- 105.Pepe M.S., Etzioni R., Feng Z., Potter J.D., Thompson M.L., Thornquist M. Phases of biomarker development for early detection of cancer. J Natl Canc Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 106.Singal A.G., Hoshida Y., Pinato D.J., Marrero J., Nault J.C., Paradis V. International liver cancer association (ILCA) white paper on biomarker development for hepatocellular carcinoma. Gastroenterology. 2021 doi: 10.1053/j.gastro.2021.01.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nass S., Moses H. 2007. Cancer Biomarkers: The Promises and Challenges of Improving Diagnosis and Treatment. [Google Scholar]
- 108.Wholley D. The biomarkers consortium. Nat Rev Drug Discov. 2014;13:791–792. doi: 10.1038/nrd4439. [DOI] [PubMed] [Google Scholar]
- 109.Bossuyt P.M., Reitsma J.B., Linnet K., Moons K.G. Beyond diagnostic accuracy: the clinical utility of diagnostic tests. Clin Chem. 2012;58:1636–1643. doi: 10.1373/clinchem.2012.182576. [DOI] [PubMed] [Google Scholar]
- 110.Chalasani N.P., Ramasubramanian T.S., Bhattacharya A., Olson M.C., Edwards V.D.K., Roberts L.R. A novel blood-based panel of methylated DNA and protein markers for detection of early-stage hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2020 doi: 10.1016/j.cgh.2020.08.065. [DOI] [PubMed] [Google Scholar]
- 111.Tao K., Bian Z., Zhang Q., Guo X., Yin C., Wang Y. Machine learning-based genome-wide interrogation of somatic copy number aberrations in circulating tumor DNA for early detection of hepatocellular carcinoma. EBioMedicine. 2020;56:102811. doi: 10.1016/j.ebiom.2020.102811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.