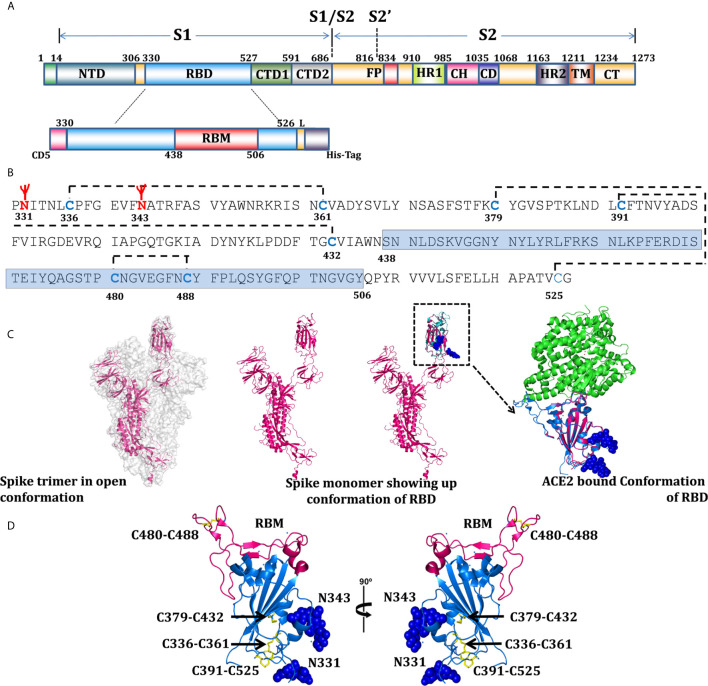
Figure 1.
Domain organization of SARS-CoV-2, RBD expression construct, and overall structural topology. (A) The schematic presentation of domain organization for the SARS-CoV-2 spike protein, differentially colored boxes represent domain boundaries; NTD (N-terminal domain), RBD (receptor binding domain), CTD1 and 2 (C-terminal domain 1 and 2), FP (fusion peptide), HR1 (heptad repeat 1), CH (central helix), CD (connector domain), HR2 (heptad repeat 2), TM (transmembrane domain), and CT (C-terminal domain) (30). The schematic representation of RBD; with N-terminal CD5 secretary sequences, coding sequences (330-526), RBM (receptor binding motif), 4 amino acid linkers, and 6x histidine tags. (B) Analysis of RBD coding sequences showing probable glycosylation sites at N331 and N343, cysteine residues - disulfide bonding network and sequences coding for RBM. (C) Surface representation of spike trimer in open conformation (6VYB) (31) with one monomeric subunit shown in the cartoon presentation to represent the overall organization of a monomer in the trimeric spike structure. One monomer of the trimer superimposed with RBD structure (6VW1) (32) and inset showing RBD in ACE2-bound conformation, showing the potential glycan in sphere representations. Residues 330-334 represented through modeled and superimposed structure (pink), showing the probable position of glycan at N331. (D) Overall structure of RBD in two orientations, RBD’s (blue), RBM shown in light pink color, cysteine residue forming disulfide bonds drawn as sticks and shown in yellow color, and two glycans are shown with sphere representation.